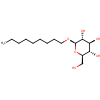
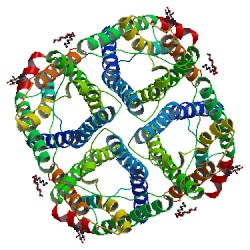
|
The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution
|
1YMG
|
|
ENTRY 1YMG SUPERSEDES
1TM8
Primary Citation
|
Biological assembly 1 assigned by authors and generated by PISA,PQS (software)
Downloadable viewers:
|