|
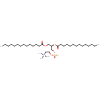
electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state
|
2B6O
|
|
Primary Citation
|
Biological assembly 1 assigned by authors
Downloadable viewers:
|