Advanced Biochemistry Fall Term, 1997
 GroE Structures and Architecture
GroE Structures and Architecture
A GroES dome caps the GroEL
"double-donut" to become a
protein folding machine
powered by ATP hydrolysis.
The pictures on this page are intended to complement the overview article by Lorimer
& Todd, "GroE structures galore" Nature Struct. Biol.3,
116-121 (1996). The material is organized in three sections:
- Two images of GroEL Structures.
Four images of GroES Structures.
GroEL & GroES Structure References list.
In addition, the following PDB files (also available in the Adv. Biochem. volume of the
BioServer) can be used to view GroEL in RasMac. To view these structures directly from
this page, click on the linked file name.
- 1grl.pdb (351 KB) One GroEL subunit as described in the Braig et al. (1994) paper.
1oelSubA.pdb (329 KB) One GroEL subunit from the refined structure described in Braig et al. (1995). This file was truncated from the original PDB coordinate file (pdb1oel.ent) to include only subunit A and its associated water molecules. The complete coordinate file (2,250 KB) is too large for convenient viewing here.
GroEL Structures
Image 1: Top and side views of the GroEL heptamer. (from
1oel.pdb)
1. The colored subunit on the left shows the crystallographic B-factor increasing from
blue shades (B=5) to red (B=150) using RasMac "Temperature" coloring.
2. The colored subunit on the right shows the domain organization of GroEL:
Note the correspondence between the B-factor coloring on the left and the domain
organization shown on the right. Click on domain organization of
GroEL for a larger picture of this subunit. It is a cartoon display made from
1oelSubA.pdb; the view is similar to that shown in Fig. 4 of Lorimer & Todd (1996).
Image 2: Top view of the GroEL ring. (from 1oel.pdb)
Starting at the top of the picture, subunits 1 & 2 are shown in the
"Domain" and "Temperature" coloring explained in Image 1. The
remaining five subunits, displayed as wireframe, show the location of residues involved in
various binding interactions.The amino acid residues are displayed as spacefill and are
colored as follows:
- Subunit 1: Cartoon display of Domains.
- Subunit 2: Cartoon display of crystallographic B-factors.
- Subunit 3: Residues involved in ATP binding (Orange), in the equatorial domain. From Fig. 3 of Boisvert, et al. (1996).
- Subunit 4: Residues (mostly hydrophobic) involved in polypeptide binding (Red), in the apical domain. From Fig. 3 of the mutational studies by Fenton, et al. (1994).
- Subunit 5: Residues involved in GroES binding (Green), in the apical domain. From Fig. 3 of the mutational studies by Fenton, et al. (1994).
- Subunit 6: Residues involved in binding the second GroEL ring (Red), at the bottom surface of the equatorial domain. From Table 2 of Braig, et al. (1994).
- Subunit 7: Residues involved in contacts between GroEL subunits within each ring. From Table 2 of Braig, et al. (1994).
Cyan: Leftside contacts (to subunit 6 in this picture).
Yellow: Rightside contacts (to subunit 1 in this picture).
GroES Structures
GroES subunit structure.
Fig. 1 from Lorimer & Todd (1996). The mobile loop is shown in orange; the 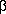 -barrel core is shown
in white; and the
-barrel core is shown
in white; and the 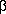 -hairpin
roof appendage is shown in red. See their caption for additional details.
-hairpin
roof appendage is shown in red. See their caption for additional details.
Top View of the GroES dome architecture.
Fig. 2 from Lorimer & Todd (1996). Same coloring scheme described above. See their
caption for additional details.
Orthogonal views of the GroES subunit structure.
Fig. 2 from Hunt et al. (1996); the mobile loop is resolved in only one of the
seven subunits. The linking segments between that 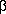 -hairpin and the
-hairpin and the 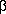 -barrel core structure are shown in yellow. The
other linking segments between
-barrel core structure are shown in yellow. The
other linking segments between 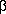 -strands are shown in cyan. See their caption for additional
details.
-strands are shown in cyan. See their caption for additional
details.
Side (a) and top (b) views of the GroES dome.
Fig. 3 from Hunt et al. (1996); same coloring scheme described above.
Structure References
- Lorimer, G.H. & Todd, M.J. (1996) "GroE structures galore." Nature Struct. Biology 3, 116.
- Braig, K. et al. (1994) "The crystal structure of the bacterial chaperonin GroEL at 2.8
 " Nature 371, 578. (ID: 1grl)
" Nature 371, 578. (ID: 1grl) - Braig, K. et al. (1995) "Conformational variability in the refined structure of the chaperonin GroEL at 2.8
 resolution." Nature Struct. Biology 2, 1083. (ID: 1oel)
resolution." Nature Struct. Biology 2, 1083. (ID: 1oel) - Boisvert, D.C. et al. (1996) "The 2.4
 crystal structure of the bacterial chaperonin GroEL complexed with ATP
crystal structure of the bacterial chaperonin GroEL complexed with ATP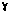 S." Nature Struct. Biology 3, 170. (ID: 1der)
S." Nature Struct. Biology 3, 170. (ID: 1der) - Hunt, J.F. et al. (1996) "The crystal structure of the GroES co-chaperonin at 2.8
 resolution." Nature 379, 37.
resolution." Nature 379, 37. - Fenton, W.A. et al. (1994) "Residues in chaperonin GroEL required for polypeptide binding and release." Nature 371, 614.
 Return to ABC97 Home Page
Return to ABC97 Home Page
Last modified November 11, 1996.
![]() -barrel core is shown
in white; and the
-barrel core is shown
in white; and the ![]() -hairpin
roof appendage is shown in red. See their caption for additional details.
-hairpin
roof appendage is shown in red. See their caption for additional details. ![]() -hairpin and the
-hairpin and the ![]() -barrel core structure are shown in yellow. The
other linking segments between
-barrel core structure are shown in yellow. The
other linking segments between ![]() -strands are shown in cyan. See their caption for additional
details.
-strands are shown in cyan. See their caption for additional
details.
Return to ABC97 Home Page