| Proteasomes: Cellular machinery for protein turnover
|
Protein degradation is a complex process that is critical to cellular regulation. In general, unlike DNA, damaged pro-teins are not repaired, but degraded, and the amino acids are recycled. In some cases, proteins age passively by gradual denaturation processes at body temperature. Proteins may also be modified by reaction with intracellular carbonyl compounds, such as glycolytic intermediates, or by reaction with products of lipid peroxidation during oxidative stress. Some proteins are induced at specific times in the cell cycle or in response to hormones, and are actively degraded as part of a cell cycle or counter-regulatory response. Some of these proteins have characteristic amino acid sequences or N-terminal residues that promote their turnover. The PEST (ProGluSerThr) sequence marks some proteins for rapid turnover, while proteins with N-terminal arginine residues generally have short half-lives. are recycled. In some cases, proteins age passively by gradual denaturation processes at body temperature. Proteins may also be modified by reaction with intracellular carbonyl compounds, such as glycolytic intermediates, or by reaction with products of lipid peroxidation during oxidative stress. Some proteins are induced at specific times in the cell cycle or in response to hormones, and are actively degraded as part of a cell cycle or counter-regulatory response. Some of these proteins have characteristic amino acid sequences or N-terminal residues that promote their turnover. The PEST (ProGluSerThr) sequence marks some proteins for rapid turnover, while proteins with N-terminal arginine residues generally have short half-lives.
|
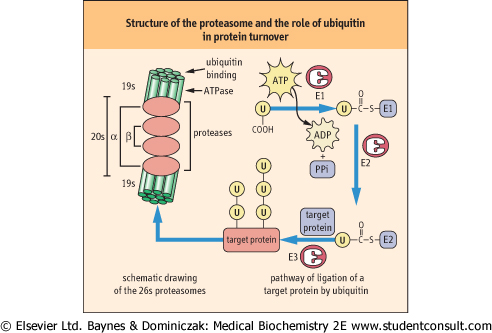
Since protein degradation is a destructive process it must be sequestered within specific cellular organelles. Lysosomes, for example, ingest and degrade damaged mitochondria and other membranous organelles. However, most soluble, cytoplasmic proteins are degraded in structures called proteasomes. The 26S proteasome consists of two types of subunits, a 20S multimeric, multicatalytic protease (MCP) and a 19S ATPase. The proteasome is a barrel-shaped structure, formed by a stack of four rings of seven homologous monomers, alpha-type subunits in the outer ends and beta-type subunits on the inner rings of the barrel. The proteolytic activity - three different types of threonine proteases - resides on beta subunits with active sites facing the inside of the barrel, thereby protecting cytoplasmic proteins from random degradation. The ATPase subunits are attached at either end of the barrel and act as gate-keepers, allowing only proteins destined for destruction to enter the barrel. The proteins are unfolded and degraded to small peptides, 6-9 amino acids in length that are released into the cytoplasm for further degradation. in length that are released into the cytoplasm for further degradation.
|
|
| Figure 32.10 Structure of the proteasome and the role of ubiquitin (u) in protein turnover. The proteasome is shown at the left, a barrel-shaped structure. The 20s multicatalytic protease in the middle rings of the barrel has protease activity on their inner face. The 19S caps at the end of the barrel have ubiquitin-binding and release and ATPase activity, and control access of proteins to the inside of the barrel for degradation. The ubiquitin cycle is involved in marking proteins for degradation by the proteasome. Ubiquitin is first activated as a thioester derivative of ubiquitin-activating enzyme E1; it is then transferred to ubiquitin-conjugating enzyme E2, then to a lysine residue on the target protein, catalyzed by ubiquitin-ligase E3. Ubiquitin is frequently co-polymerized on target proteins. The more highly ubiquinated the protein, the more susceptible it is to proteasomal degradation. Note that the drawing is not to scale; the proteasome is a 26S macromolecular complex (>2 000 000 D); target proteins are smaller while ubiquitin molecular weight is less than 10 000 D. |
| Proteins destined for destruction are directed to the proteasome primarily by covalent modification with a highly conserved, 76 amino acid residue protein called ubiquitin which is found in all cells. Ubiquitin must be activated to fulfill its role (Fig. 32.10) and this is accomplished by a ubiquitin-activating
enzyme called E1. Activation occurs when E1 is attached via a thioester bond to the C-terminus of ubiquitin via ATP-driven formation of a ubiquitin-adenylate intermediate. The activated ubiquitin is then attached to a ubiquitin carrier protein, known as E2, by a thioester linkage. A ubiquitin protein ligase known as E3 transfers the ubiquitin from E2 to a target protein, forming an isopeptide bond between the carboxyl terminus of ubiquitin and the ε-amino group of a lysine residue on a target protein. Denatured and oxidized proteins are commonly ubiquitinated by this mechanism. The 26S subunit of the proteasome has a ubiquitin binding site that allows proteins with a covalently attached ubiquitin protein to enter the barrel; the ubiquitin is then released by a ubiquitinase activity and recycled to the cytosol. Polymerization of ubiquitin on target proteins (polyubiquitination) enhances degradation of the protein.
|
| page 457 |  | | page 458 |
| The ubiquitin pathway leading to proteasomal degradation of proteins is complex. There are a number of E2 and E3 proteins with differential target specificity, and six different ATPase activities associated with the 19S subunit. The ATPases are thought to be involved in denaturation of ubiquinated proteins. These variations, as well as changes in response to the cell cycle and hormonal stimulation, provide a flexible and regulated pathway for protein turnover.
|
|