|
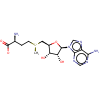
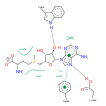
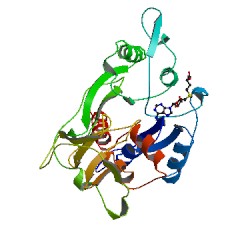
CRYSTAL STRUCTURE OF THE HHAI DNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYL-L-METHIONINE
|
1HMY
|
|
Primary Citation
|
Biological assembly 1 assigned by authors
Downloadable viewers:
|