|
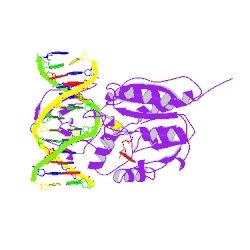
COVALENT TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE, DNA AND S-ADENOSYL-L-HOMOCYSTEINE
DOI:10.2210/pdb1mht/pdb NDB ID: PDEB08
|
1MHT
|
|
Primary Citation
|
Biological assembly 1 assigned by authors
Downloadable viewers:
|