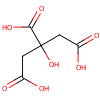
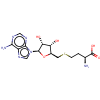
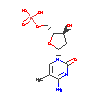
|
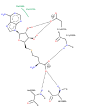
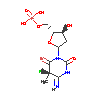
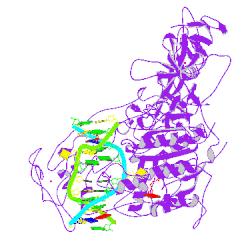
Structure of mouse DNMT1 (731-1602) bound to hemimethylated CpG DNA
DOI:10.2210/pdb4da4/pdb NDB ID: NA1539
|
4DA4
|
|
Primary Citation
|
Biological assembly 1 assigned by authors and generated by PISA (software)
Downloadable viewers:
|