|
|
|
Back to the
Index...DNA Meth Word document: MethylTrans11.doc
July 2011 Molecule of the Month by David Goodsell 2QRV 3PT6 1MHT
DNA Methyltransferases DNMT1, DNMT3, restriction modification system Hha1
doi: 10.2210/rcsb_pdb/mom_2011_7 (ePub Version )
Keywords: epigenetics, DNA methylation, DNMT1, DNMT3, restriction modification system Hha1
|
|
|
Introduction Your body is built of skin cells, nerve cells, bone cells, and many other different types of cells.
These cells are different shapes and sizes, and each type of cell builds a characteristic collection of proteins that are needed for its function. However, every cell in your body contains the same genetic information, encoded in strands of DNA. How does each cell decide which genes to use and which ones to ignore?
Genetics and Epigenetics Scientists have discovered that the information in DNA does not end at the simple genetic sequence of bases. Cells layer additional forms of control on top of the genetic code, creating "epigenetic" information that modifies the use of particular genes. In some cases, this control is performed by the positioning of nucleosomes. In other cases, bases in the DNA are methylated, modifying how they are read during protein synthesis.
Clean Slate In the first minutes of life, when we are composed of a single cell, this epigenetic information has been wiped clean. In the fertilized egg, the methyl groups have been removed and every gene is like all the others. Then, as cells divide in the embryo, they have to make choices about what they are going to do-becoming skin cells or nerve cells or their particular fate. At this point, DNA methyltransferases come into play, and they add methyl groups to genes, shutting off some and activating others. The DNA methyltransferase DNMT3, shown here from PDB entry 2QRV, performs this important job, creating the proper epigenetic coding of methyl groups throughout the genome.
The information is propagated in a tricky way. Methyl groups are almost always added to Cytosine bases with the sequence:
---CG---
---GC---
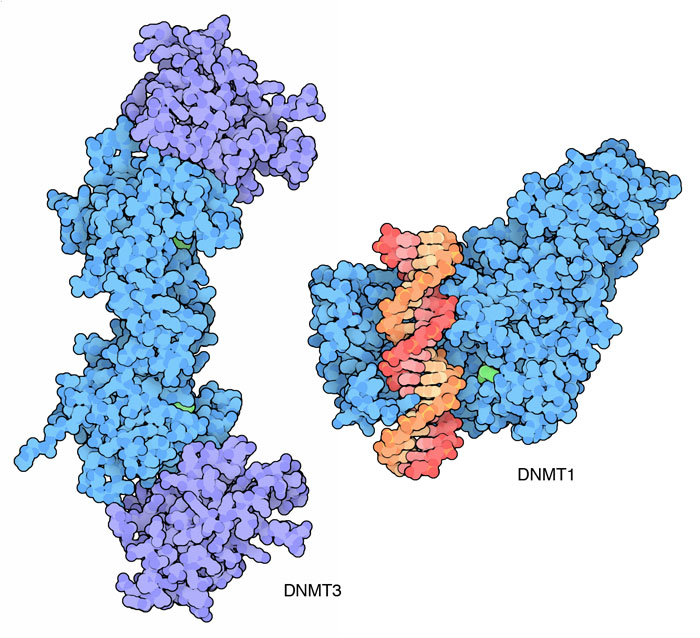
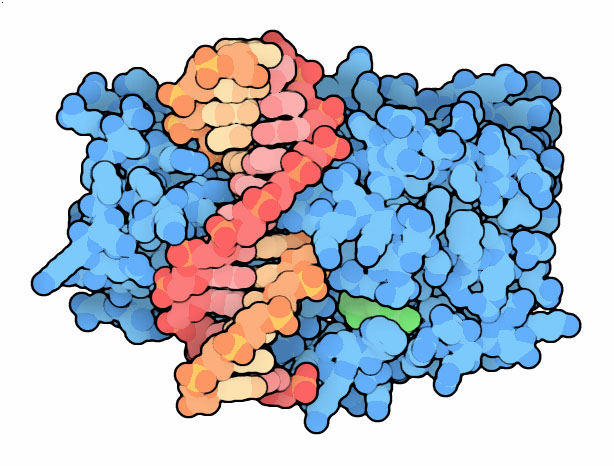
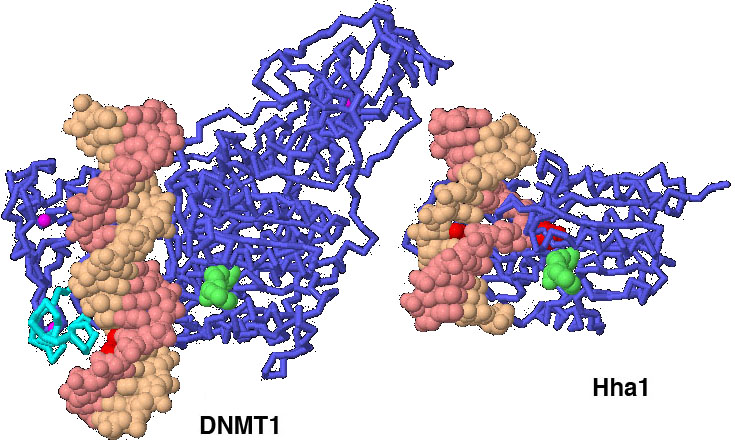
click on the image for an interactive Jmol 3PT6 1MHT
Topics for further exploration The methyl group is provided by the cofactor S-adenosylmethionine during the methylation reaction. Many of the DNA methyltransferase structures were solved with the similar cofactor
S-adenosyl homocysteine in the active site, but PDB entry 3av6 is DNMT1 with SAM bound. Can you find other enzymes in the PDB that use this cofactor? Are they performing similar reactions?
You can use the structure comparison tool to overlap structures of DNMT enzymes and bacterial methyltransferases. Notice that the cores of the enzymes are similar, but the DNMT enzymes are larger. Why do you think our DNA methyltransferases need additional protein domains along with the domain that performs the reaction?
![]() PT6
PT6
References
R. Z. Jurkowska, T. P. Jurkowski and A. Jeltsch (2011) Structure and Function of Mammalian
DNA Methyltransferases. ChemBioChem 12, 206-222. X. Cheng and R. M. Blumenthal (2008) Mammalian DNA Methyltransferases: A Structural Perspective. Structure 16, 341-350.