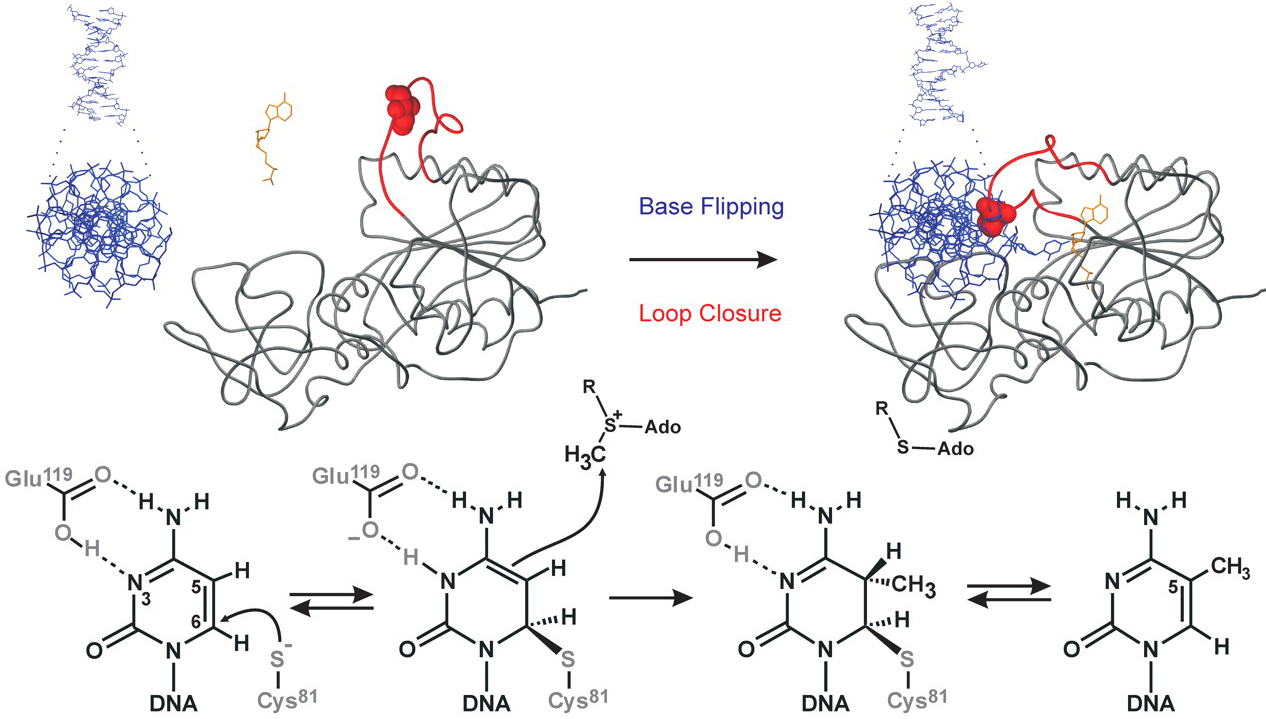
Figure 1. Conformational transitions and covalent catalysis by M.HhaI. (A) Upon binding of DNA and cofactor, M.HhaI flips its target cytosine out of the DNA helix into the active site; the catalytic loop in the protein makes a large motion to lock the target base and the bound cofactor. M.HhaI is shown as backbone trace, the catalytic loop (residues 81-100) is red, the engineered Ile86 residue is shown as space fill, DNA and cofactor are represented as sticks models in blue and orange, respectively. (B) The mechanism of covalent target base activation and Methyl group transfer by M.HhaI along with associated spectral changes of the target base. (C) General kinetic scheme of conformational transitions upon formation of an unproductive ternary complex with AdoHcy (upper) and during catalytic turnover in the presence of AdoMet (lower). C, cofactor (AdoHcy); E, enzyme; D, DNA; m, Methylgroup on cofactor or DNA; F, flipped out conformation; L, locked loop conformation; dot, non-covalent association; hyphen, a covalent bond between enzyme and DNA.Back to the methylase tutorial. 