Transcription Translation
Tentative Lecture
schedule Biochemistry
of Medicinals I
Phar 6151 Part 2
– Nucleic Acids
Instructor: Dr. Natalia Tretyakova, Ph.D. «hyperlink
"mailto:Trety001@umn.edu"»
- 6-3432
PDB reference correction
and design Dr.chem., Ph.D. Aris Kaksis, Associate Professor e-mail:
ariska@latnet.lv
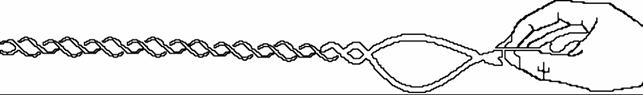
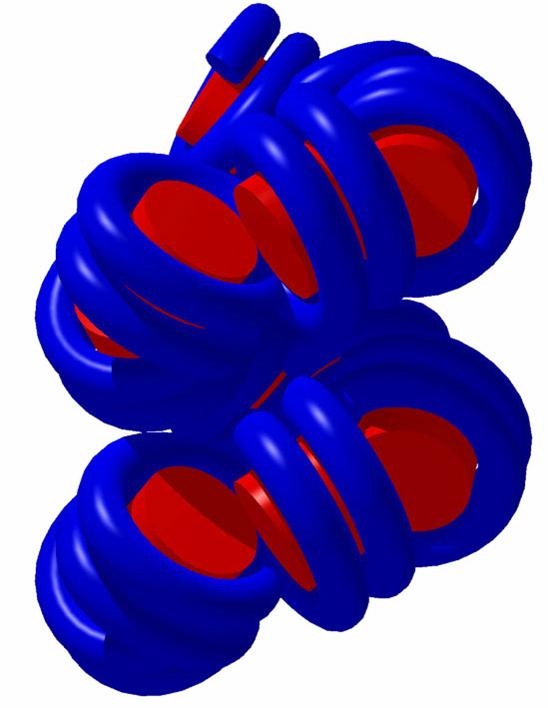
DNA Topology
1. DNA has to be coiled to fit inside
the cell - interior the nucleus
Average human chromosome: 2.05•108 nucleotides (8.75 cm) 24*8.75=210.00 23.529410*8.75=205.882338
Nucleus diameter: about 10 µm
DNA polymers have
to be folded into to fit into the nucleus (tertiary
structure 3°).
2. Many viruses and bacteria
have circular genoms – tertiary structure
3°.
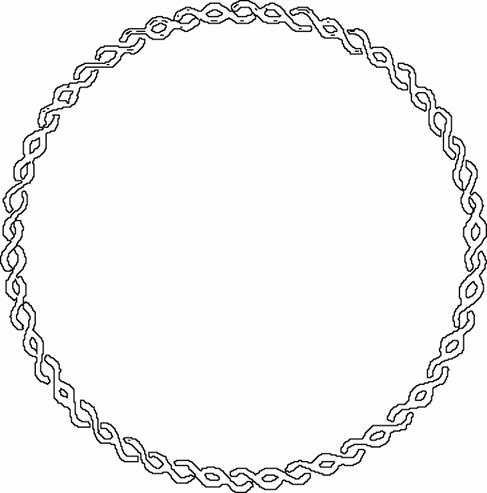
DNA Topology:
Negative
supercoiling :
DNA is twisted
in the direction opposite ¯
to the direction ¯
of
the double helix (underwound)
Positive
supercoiling :
DNA is twisted in the same direction ¯¯ as
the direction ¯
of the double
helix (overwound)
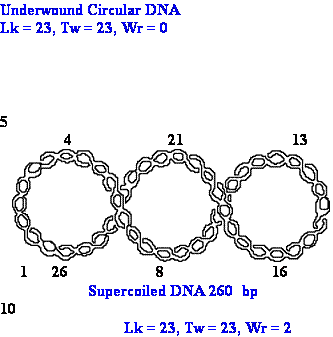
DNA Topology: linking number
Topoisomers can
be quantitatively defined by the linking
number (Lk).
Lk is the number
of times a strand of DNA winds in the
right handed direction around the helix
axis when the axis
is constrained.
Tw (twist) is the helical
winding of the strands
around each other.
Wr (Writh) is the number of superhelical turns
Lk = Tw + Wr, if Lk = const., DTw =
- DWr
DNA Topology:
example
®
1
®
5
®
10
®
15¯
®
25
¬
20
¬
15 ¯
 ¬¿
¬¿
Relaxed B-DNA 260 bp 25
1


1
®
5
®
10
®
15¯
 ¯
¯
®
20
®
23
®
25
1


DNA Topology:
chromosomal DNA
• Chromosomal DNA
is organized in loops (no free ends)
• It is negatively
supercoiled 1 (-) supercoil per 200
nucleotides
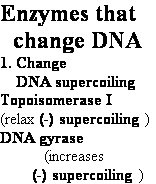
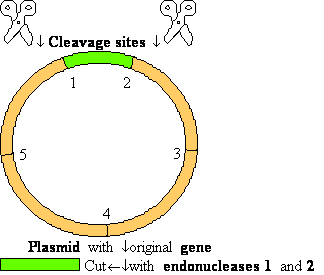
2. Cut in pieces
Exonucleases 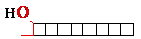 dNMP +
dNMP + 
Endonucleases 
 +
+ 
3.
Ligate DNA
- DNA ligase  +
+ 

DNA Gyrase •
Introduces negative supercoils
• Requires energy (ATP)
• Necessary for DNA synthesis

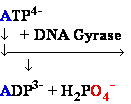

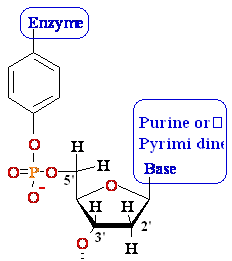 DNA Gyrase
DNA Gyrase
(+) supercoiled DNA (-)
supercoiled DNA
DNA chain
DNA containing
one 1 positive (+) SuperCoil

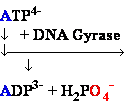
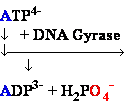
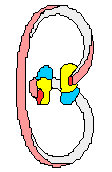
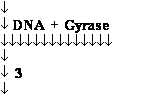

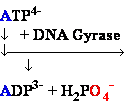
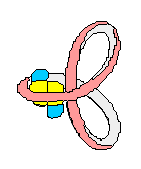
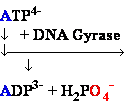
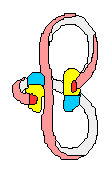
DNA containing
one 1 negative (-) SuperCoil
Drugs
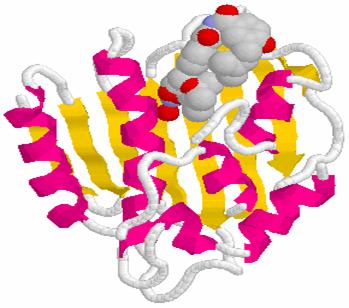
that inhibit DNA gyrase chlorobiocin
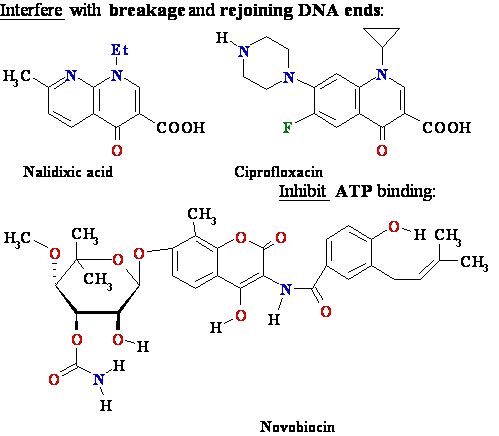
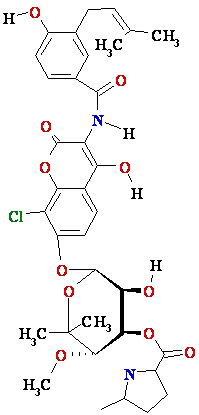
DNA Gyrase-Novobiocin
1AJ6.PDB; clorobiocin
+ Gyrase 1KZN.spt
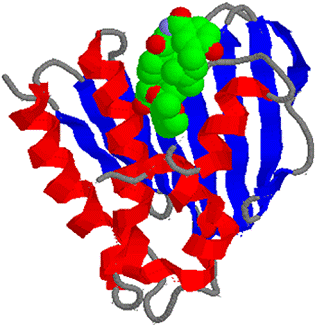
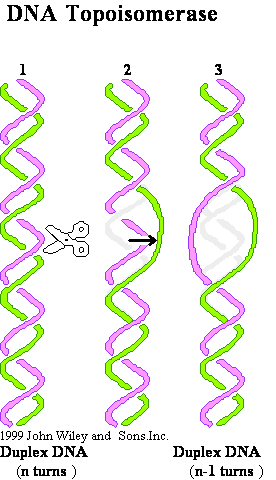
DNA Topoisomerase
Relax supercoiled DNA: W = + 1 to ® W = 0
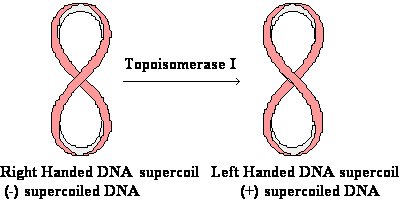

Human DNA
Topoisomerase I: 1EJ9.PDB side view
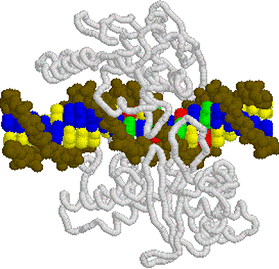
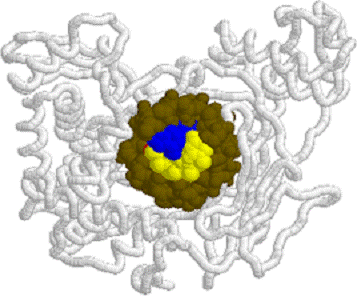
1EJ9.PDB
side view Human DNA Topoisomerase I 1EJ9.PDB
top view
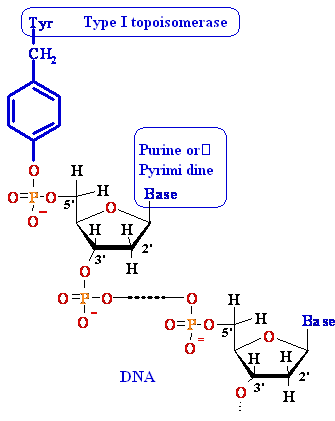
Drugs that inhibit DNA Topoisomerase I
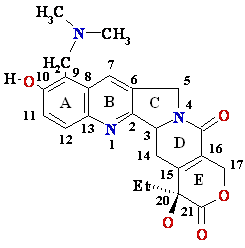 Topotecan
(Hycamtin)• Anti- tumor
activity
Topotecan
(Hycamtin)• Anti- tumor
activity
• Stabilizes topoisomerase
I-DNA intermediate, preventing DNA
strand re-ligation
Enzymes that
cut DNA: exonucleases
Exonucleases  dNMP +
dNMP + 
Endonucleases 
 +
+ 
• Degrade DNA in
a stepwise manner by removing mononucleotides in 5'
---> 3'
or 3' --->
5'
direction
• Require a free —OH
• Most exonucleases
are active on both single and double-stranded DNA
• Used for degrading foreign DNA and in proofreading
during
DNA synthesis
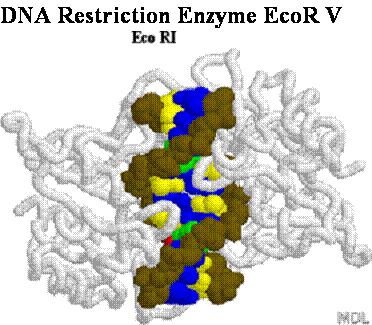
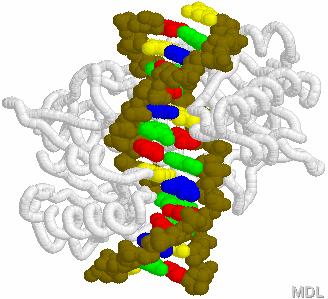
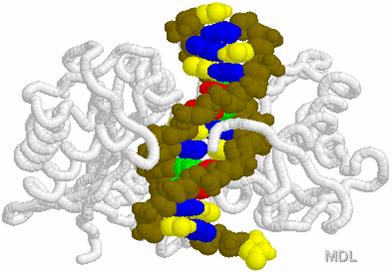
DNA Restriction Endonucleases
2.
Cut in pieces • Cleave internal phosphodiester
bonds resulting in 3'-OH and 5'-phosphate
ends
Endonucleases 
 +
+ 
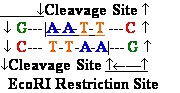
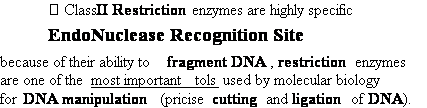
• Found in bacteria where they are used for protection
against foreign DNA
EcoRI
recognition site: Eco - for
Escherichia coli ;
Pvu - for Proteus
Vulgaris 1PVI.PDB


* asterisks - idicates bases
that are methylated (where
known) ; Bam - for Bacillus
amylolique-faciens
Biochemistry, 4th Edition, W.H.Freeman Company
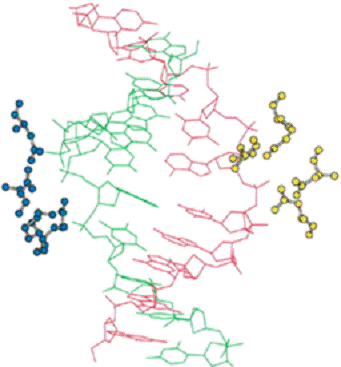
Pvu
II
Bam
H
PvuII - for 1PVI.PDB
Proteus Vulgaris Eco
- for 1RVA.PDB
Escherichia colli
Bam - for 1BHM.PDB Bacillus
amyloliquefaciens H
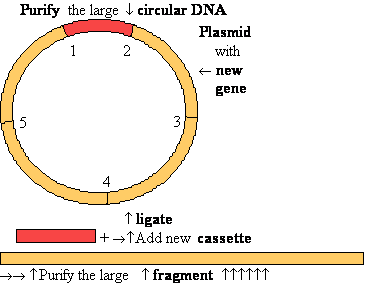
DNA Ligase Ligate DNA
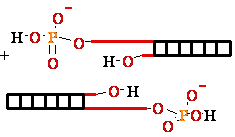


• Forms phospho-di-ester
bonds between 3'OH
and 5'
phosphate
• Requires double-stranded
DNA
• Activates 5’phosphate
to nucleophilic attack by trans-esterification with activated AMP
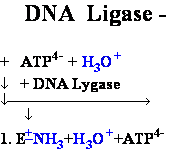 --->
--->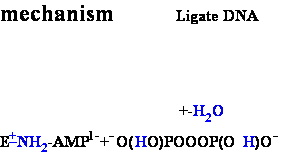
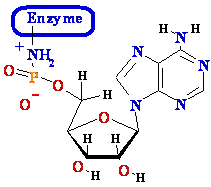
2. E-+NH2-AMP1- + H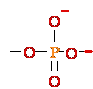 5'-DNA ---> E-+NH3 +
AMP1--
5'-DNA ---> E-+NH3 +
AMP1--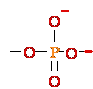 5'-DNA
5'-DNA
3. DNA-3'-OH + H2O + AMP1--O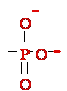 5'-DNA --->
DNA-3'--O
5'-DNA --->
DNA-3'--O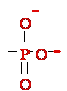 5'-DNA +H2HO+ + AMP2-
5'-DNA +H2HO+ + AMP2-
DNA cloning








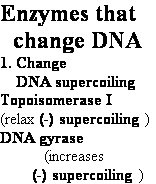

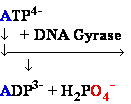

 DNA Gyrase
DNA Gyrase
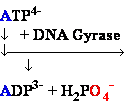
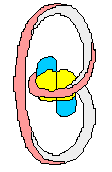
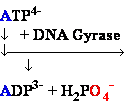
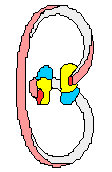
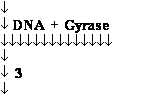

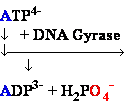

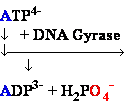
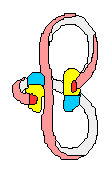




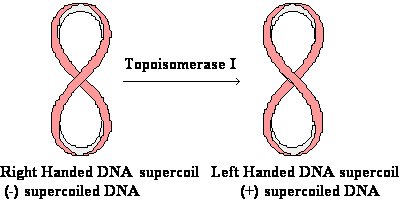





 Topotecan
(Hycamtin)• Anti- tumor
activity
Topotecan
(Hycamtin)• Anti- tumor
activity 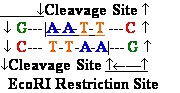
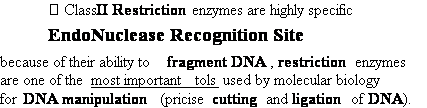








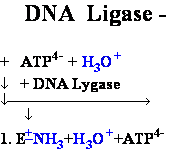 --->
--->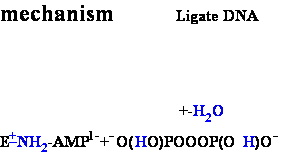

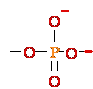 5'-DNA ---> E-+NH3 +
AMP1--
5'-DNA ---> E-+NH3 +
AMP1--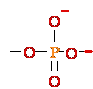 5'-DNA
5'-DNA
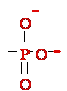 5'-DNA --->
DNA-3'--O
5'-DNA --->
DNA-3'--O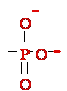 5'-DNA +H2HO+ + AMP2-
5'-DNA +H2HO+ + AMP2-
