Instructor: Dr. Natalia Tretyakova, Ph.D. «hyperlink
"mailto:Trety001@umn.edu"»
- 6-3432
PDB reference correction and design Dr.chem., Ph.D. Aris
Kaksis, Associate Prof. e-mail:
:ariska@latnet.lv
 Template
Template
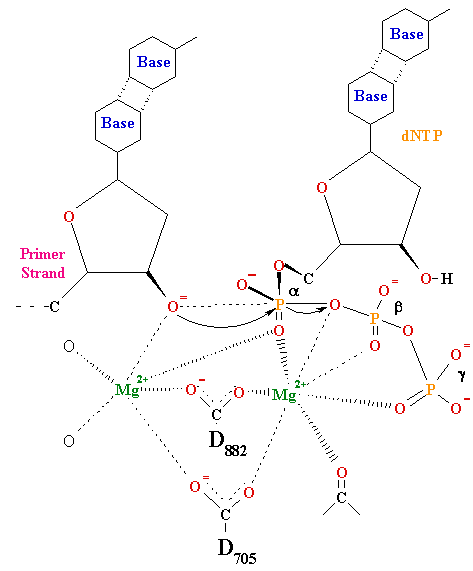
Figure 1. Two – metal – ion mechanism.
E. Coli DNA Polymerases
Characteristic
Pol
I
Pol II
Pol III
Mol. Weight (Da)
103,000
88,000
900,000
Number of polypeptides
1
4
10
rate (nucleotides/sec)
16-20
7
250-1000
3'--> 5' exonuclease
yes
yes
yes
5'--> 3' exonuclease
yes
no
no
function
Primer
removal, unknown
Major
gap filling
replicative
polymerase
DNA
Polymerization Has Three Stages
1) Initiation
2) Priming
3) Processive Synthesis
DNA Polymerization:
Initiation
•
DNA replication begins at a specific site.
•
Example: oriC site
from E. coli.
• 245 bp out of 4,000,000 bp
• contains tandem array of three 13-mers; GA C
C N
N
 N
N



• GA C common motif in oriC
C common motif in oriC
• A bp
are common to facilitate unwinding
bp
are common to facilitate unwinding
Initiation of
DNA Polymerization
Tandem Array
(13-Mer Seq. A- Rich)
dna
A
Rich)
dna
A
Ori C
dna A
protein unwinds oriC at
several sites
Enzymes
involved in the initiation of DNA
Polymerization
Enzyme
Function
dna A
recognize replication origin and
unwinds
it at several sites
Helicase (dnaB)
unwinding of ds DNA
DNA gyrase
generates (-) supercoiling
that compensates for the (+)
supercoiling generated in front of the replication fork
SSB
stabilize unwound ssDNA
Primase
a RNA
polymerase that generates RNA primer.
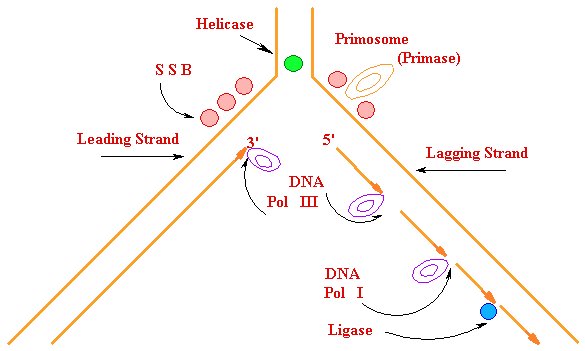
Priming of DNA
Polymerization Primer
Synthesis and removal
3'  5'
5'
|| Primase
3'  5'
5'
5'  3'
3'
RNA primer  + || DNA Pol
III
+ || DNA Pol
III
3'  5'
5'
5' 
 ----> 3'
----> 3'
|| DNA Pol I
DNA strand
3'
 5'
5'
5'  3'
3'
degraded 






 <-- primer + || DNA
Ligase
<-- primer + || DNA
Ligase
3'  5'
5'
5'  3'
3'
DNA Polymerization
Processive Synthesis
The synthesis of DNA in E. Coli is carried out by the DNA
polymerase III holoenzyme.
• Extremely
high processivity; once it combines with the DNA and starts it
does
not come off until finished.
• Tremendous
catalytic potential; 1000 nucleotides per sec.
• Low error
rate (high fidelity) 1 error per 10,000,000,000
alpha subunit is the polymerase
epsilon subunit is the 3'--> 5' exonuclease
beta subunit provides a tight clamp for
sliding DNA through
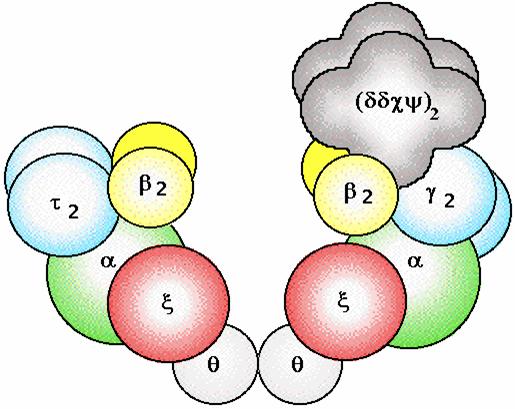 Pol III
Pol III
Pol III
Pol III + DNA
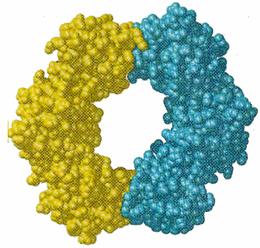
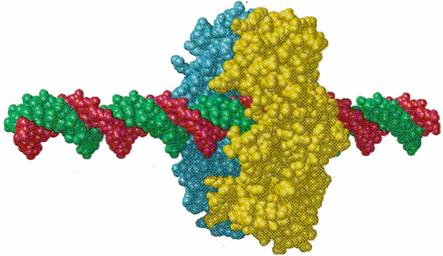
½¾
50 Å ¾
¾ ½
DNA
SynthesisDNA Synthesis
5' 3'
3' 5'
3'
5'
 Template
Template 
 Pol III
Pol III


