Instructor: Dr. Natalia Tretyakova, Ph.D. «hyperlink
"mailto:Trety001@umn.edu"»
- 6-3432
PDB reference correction and design Dr.chem., Ph.D. Aris
Kaksis, Associate Prof. e-mail:
:ariska@latnet.lv
Fidelity
of Polymerization: Absolutely Essential !!
Error
Probability = Polymerization error 10-4
X
3'--> 5'
Nuclease error
10-3
=
10-7 or 1 in 10,000,000
DNA Synthesis: addition of
new dNTPs follows Watson-Crick rules
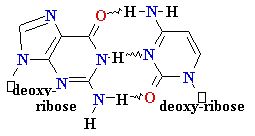
G = C
A=
Polymerase
errors
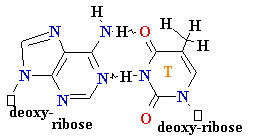
• Very low rate of
misincorporation (1 per 108)
• Errors can occur due to the
presence of minor
tautomers of nucleobases.
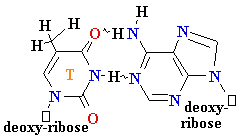
 = A
Cytosine C = A Rare
tautomer of A
= A
Cytosine C = A Rare
tautomer of A
Normal base pairing
Mispairing
Types
of DNA Mutations
1.
Point mutations:
substitution of one base
pair for another, e.g. A for GC
for GC
• the most common form of mutation
• transition;
purine to purine and
pyrimidine to pyrimidine
•transversions; purine to pyrimidine or
pyrimidine to purine
2.
Deletion of one or
more base pairs
3.
Insertion of
one or more base pairs
Termination of Polymerization:
The Key to Nucleoside
Drugs
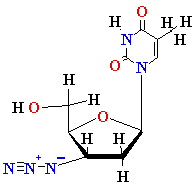
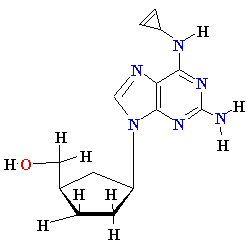
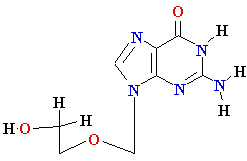
AZT
Ziagen
Acyclovir
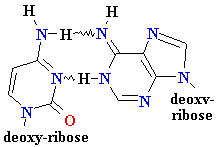
Normal base pairing in DNA
and
an example of mispairing via chemically modified nucleobase
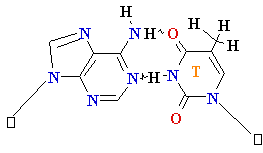
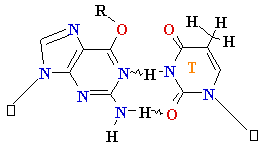
Adenine A=
 O6-Alkyl-Guuanine
O6-Alkyl-Guuanine 
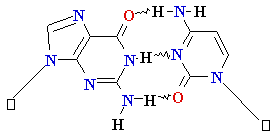
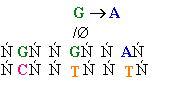
Guanine
G = C Cytosine
Chemical Mutagens
Mutations can occur
when the normal
bases that are incorporated are changed.
1.
Base analogs or bases that have altered hydrogen bonding capabilities
can cause
transitions.
Ex. Bromo and Guanine
or 2-aminopurine and cytosine
and Guanine
or 2-aminopurine and cytosine
2.
Bases can be modified on the DNA by mutagens.
•adenine
is
oxidatively deaminated to
hypoxanthine, cytosine
to  , guanine
to xanthine
, guanine
to xanthine
3.
Intercalating Agents • insertion and
deletion mutants
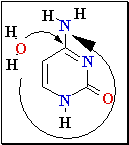
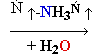
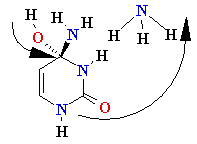
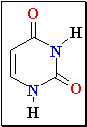 Cytosine Deamination
Cytosine Deamination
DNA
Damage
Base pairs can be damaged by
environment or chemical
factors, such as UV light or nitrosureas
.
1. Pyrimidine
dimers due to UV light
• repaired by excision of the region by uvrABC
excinuclease followed by gap filling by DNA Pol I.
•
can be repaired by photolyase, which
splits the dimers
•
cause of xeroderma pigmentosum, don't
have excinuclease
2. Nitrosoureas
and
other alkylating agents are obtained from smoking and other chemical
contaminants.
• result in methylation
of G to form O6Methyl
G,
which base pairs with T and T to form
O4Methyl
T, which base pairs with G
there are some demethylation enzymes
3. Cytosine
deamination to Uracil
• spontaneous
• uracil-DNA
glycosidase, AP exonuclease, DNA Pol I.
4. DNA repair
is based on methylation
of parental strand and a mismatch
repair system that
replaces
the incorrectly incorporated base.
DNA
Synthesis: Take Home Message
1) DNA synthesis is
carried out by DNA polymerases with high
fidelity.
2) DNA
synthesis is characterized by initiation,
priming and processive synthesis steps
and proceeds in 5’ --> 3’
direction.
3) Modifications
of DNA base pairs, if not repaired,
can lead to mutations
of
the DNA sequence.











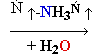
 Cytosine Deamination
Cytosine Deamination
