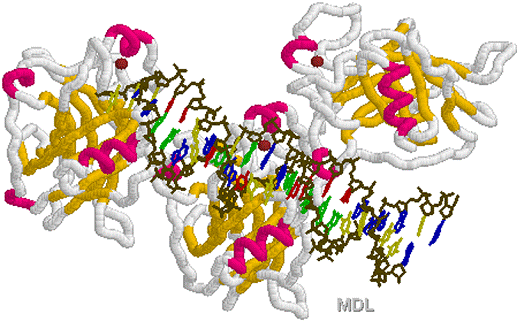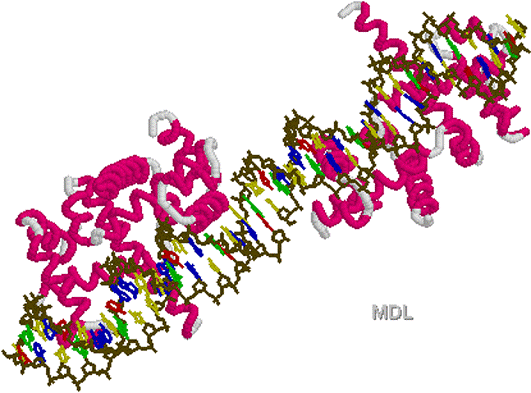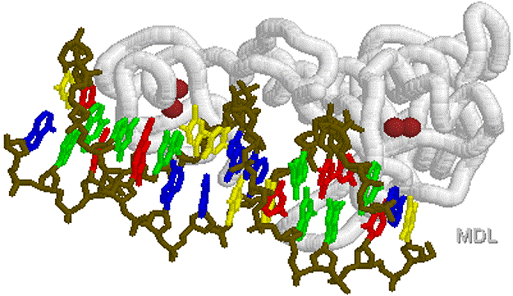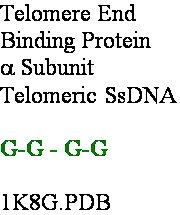
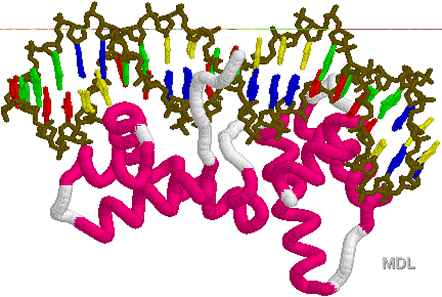
 Helix
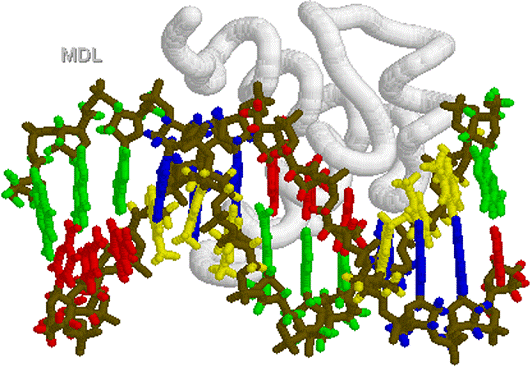
loop turn Helix
Helix
loop turn Helix 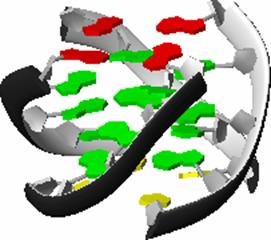
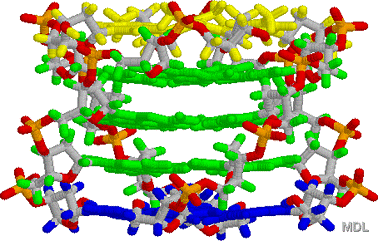
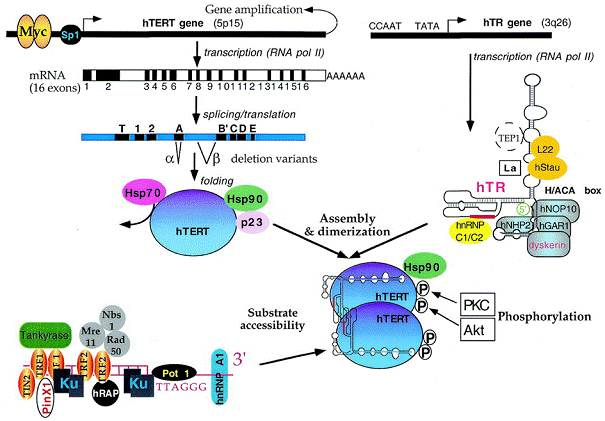
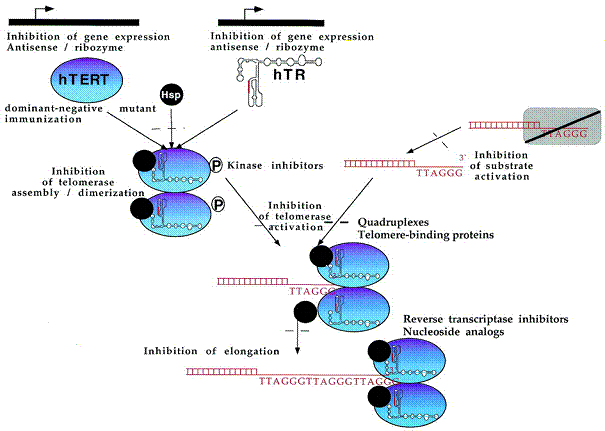
 2
2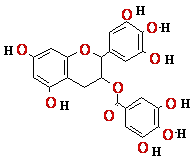
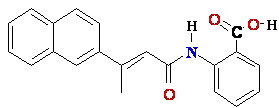 4
4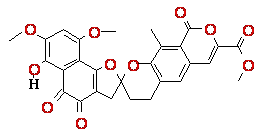 5
5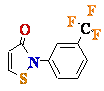
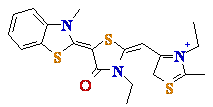 7
7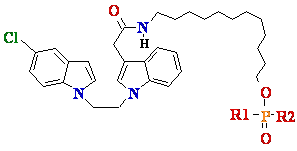 8
8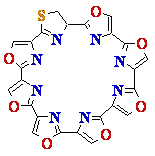
| Familya |
IC50b (µM) |
Targetc |
Cell effectd |
| BIBR
1532 |
0.093 | hTERT |
D |
|
ß-Rubromycin |
3 | ? |
? |
|
Isothiazolones (TMPI) |
1 | hTERT? |
? |
|
Rhodacyanines (FJ5002) |
2 | hTERT? |
? |
|
Bis-indoles |
2 | ? |
? |
|
Catechins (EGCG) |
1 | ? |
? |
|
Telomestatin |
0.005 | ? |
? |
|
TDG-TP |
0.06 | Nucleoside |
? |
|
Ribozymes |
? | hTR |
I |
|
PNA |
<0.001 | hTR |
D |
|
2'-OMe (oligonucleotide) |
hTR | D |
(238,239) |
|
2'-MOE (oligonucleotide) |
0.005 | hTR |
D |
|
2'-5'A-oligonucleotide |
? | hTR |
I/D |
|
N3'P5' phosphoramidates |
<0.001 | hTR |
? |
|
Dibenzophenanthrolines |
0.03 | G4 |
? |
|
Acridines |
0.06 | G4 |
? |
|
RHPS4 (pentacyclic
acridine) |
0.3 | G4 |
D |
|
Ethidium |
0.03 | G4 |
? |
|
Triazines |
0.04 | G4 |
D |
| Bis-acridine
|
0.75 | G4 |
? |
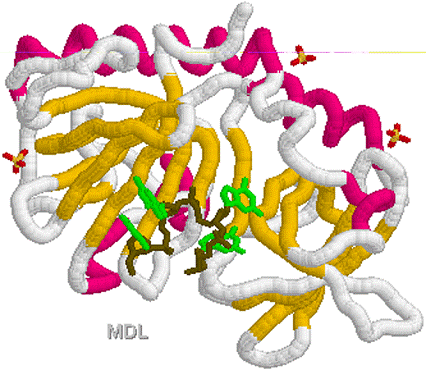
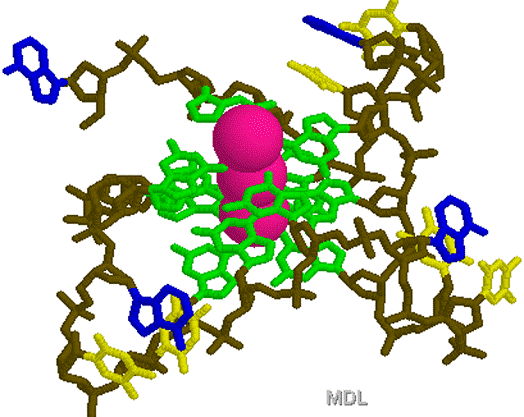
 1KF1.PDB
1KF1.PDB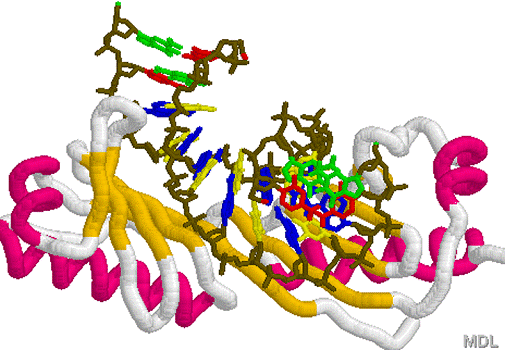 1TGH.PDB
1TGH.PDB