
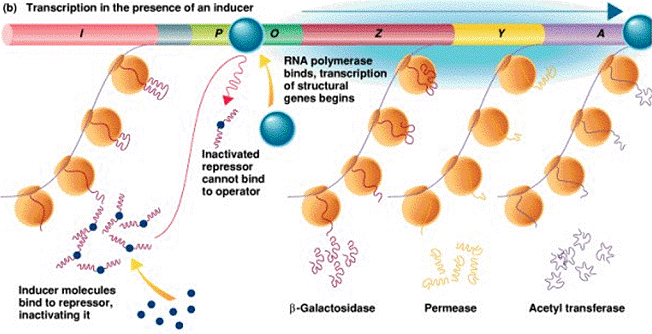
| Prokaryotes: 1) Primary transcript is translated directly (no splicing) 2) mRNA are often «polycistronic» (i.e. encode more than one 1 polypeptide) |
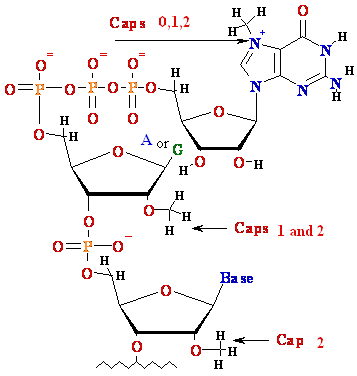
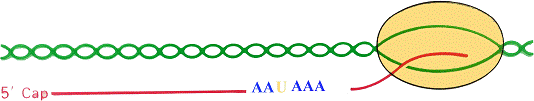
Eukaryotes: 1) Primary transcript is processed - capped, PolyA added, spliced 2) mRNA are transported to cytoplasm 3) Each mature mRNA codes for one 1 polypeptide |

 3'
5'
3'
5'  3'
¾®5'
3'
¾®5'  3'
3' 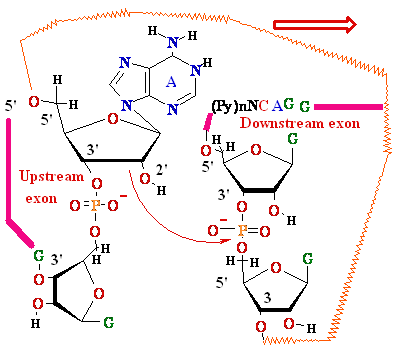
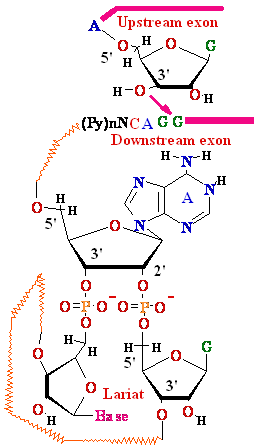
 3'
3' 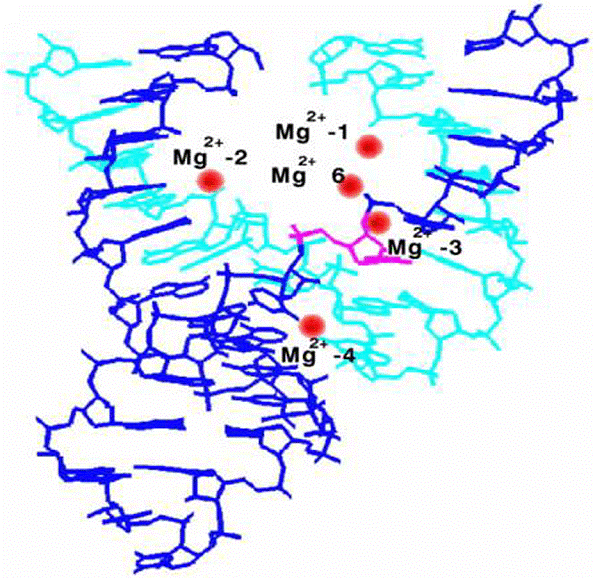
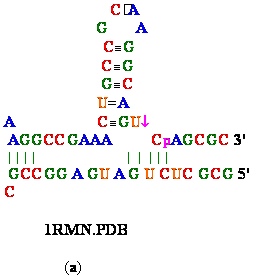
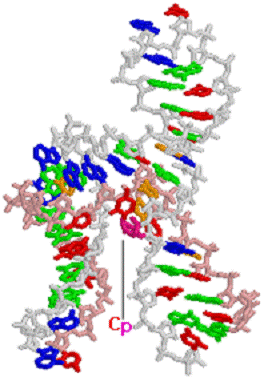 (b)
(b)