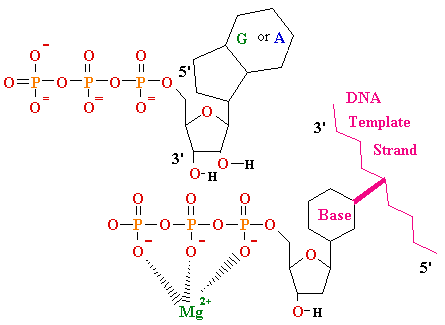
| DNA |
RNA |
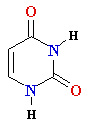
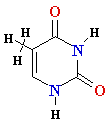 Thymine T Thymine T 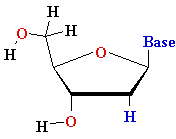 2'-de-oxy-ribose 2'-de-oxy-ribose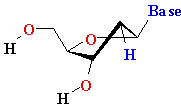 |
 Uracyl U Uracyl U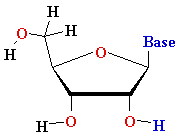 D-ribose D-ribose 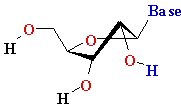 |
| DNA |
RNA |
 Thymine T Thymine T  2'-de-oxy-ribose 2'-de-oxy-ribose |
 Uracyl U Uracyl U D-ribose D-ribose  |
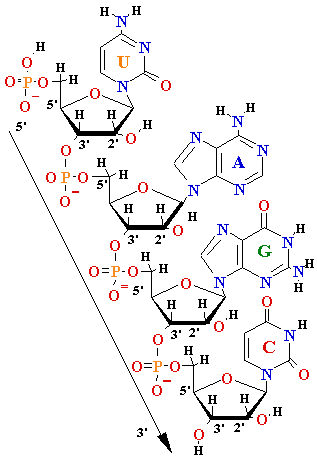
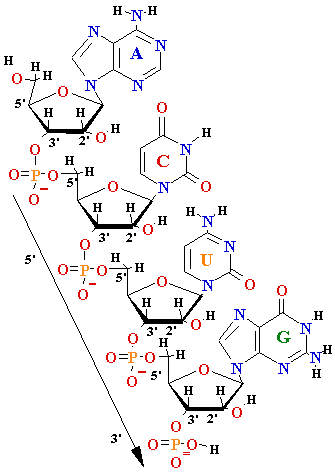






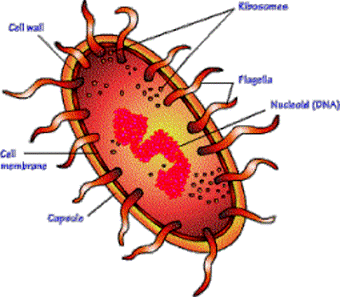
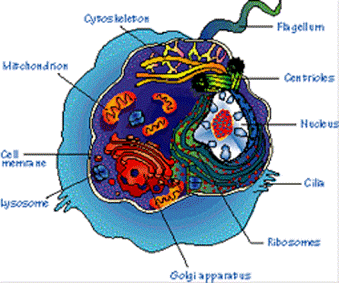
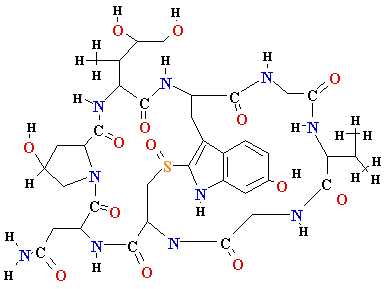

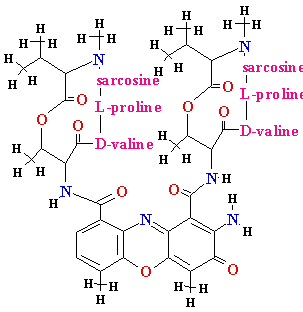
| Gene |
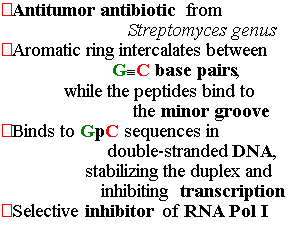
Protein Mass(kDa) |
Deletion Mutant |
E.Coli analog |
| RPB1 RPB2 RPB3 RPB4 RPB5 RPB6 RPB7 RPB8 RPB9 RPB10 RPB11 RPB12 |
190 140 35 25 25 18 19 17 14 8 14 7.7 |
nonviable nonviable nonviable nonviable conditional nonviable nonviable nonviable conditional nonviable nonviable nonviable |
b b a - - - s |
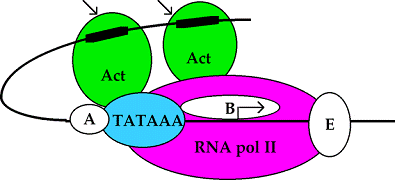
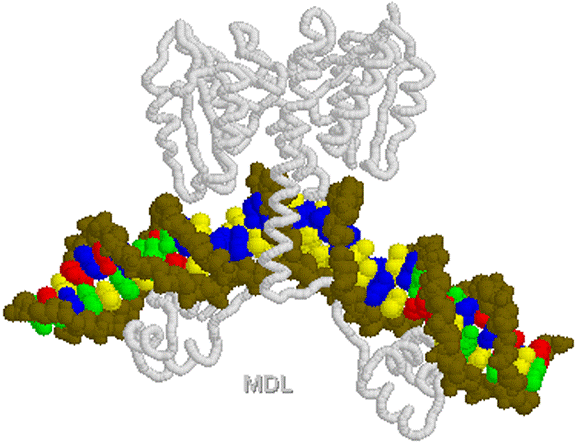
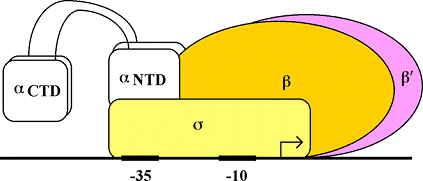
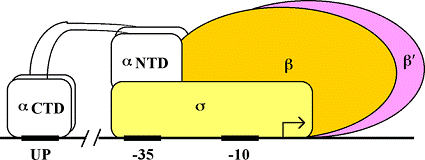
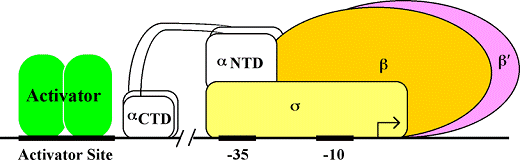
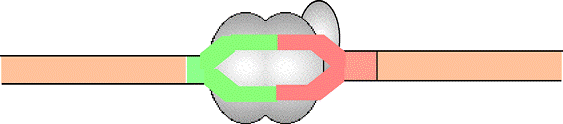
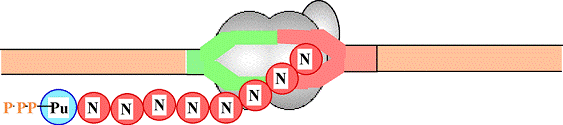
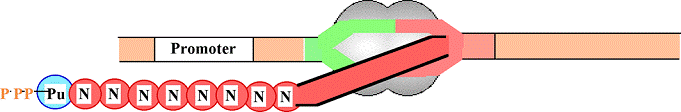
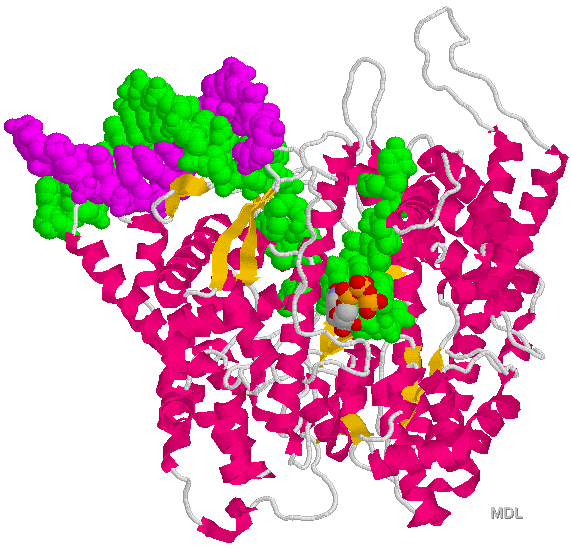
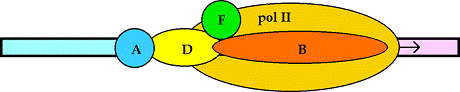 Pol II binds IIB
Pol II binds IIB
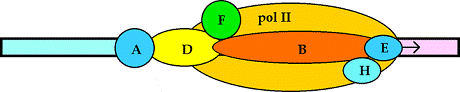

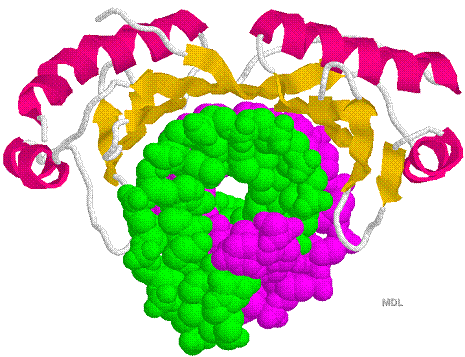
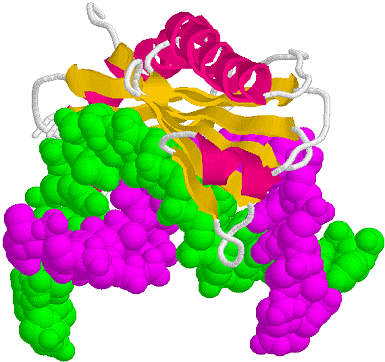
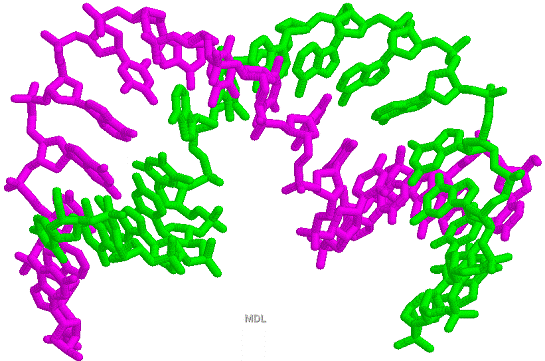
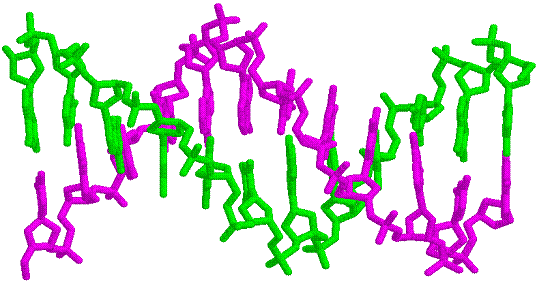
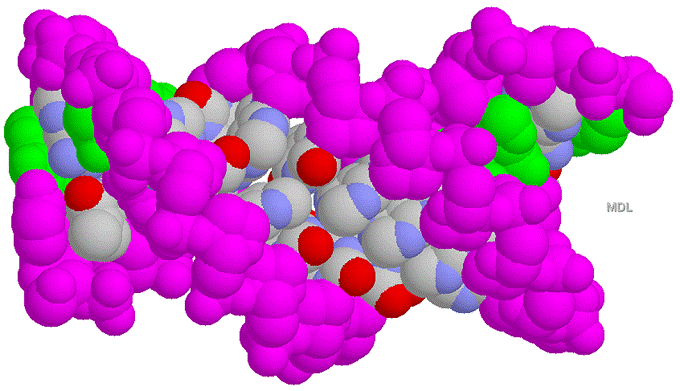
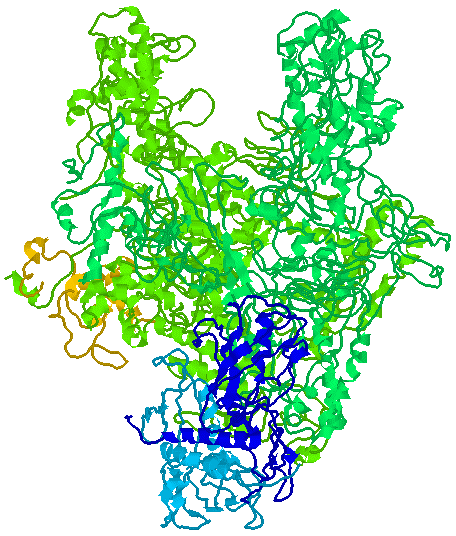
 Figure 33-28 page 856
Figure 33-28 page 856
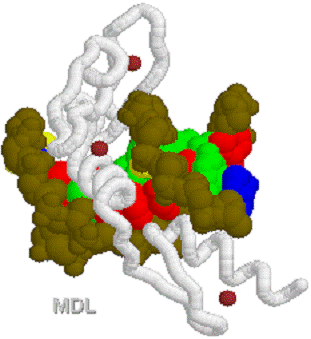

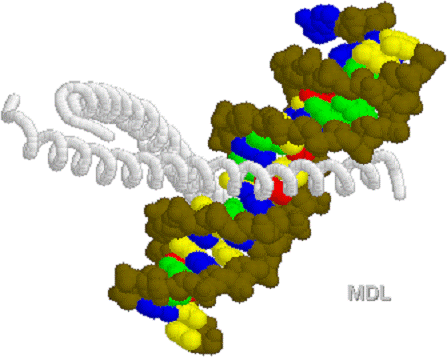
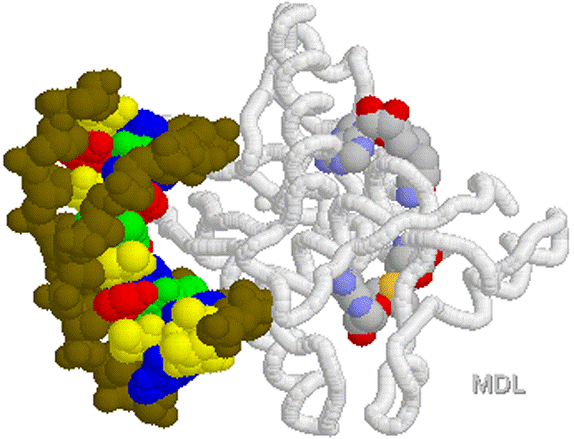
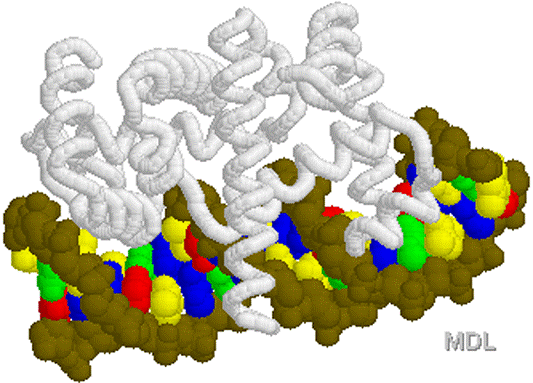
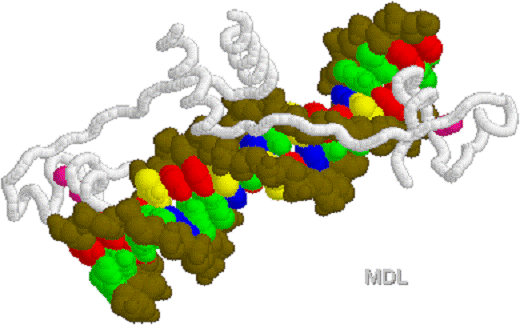 1D66.PDB-Zn2+-Gal4
1D66.PDB-Zn2+-Gal4