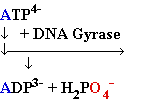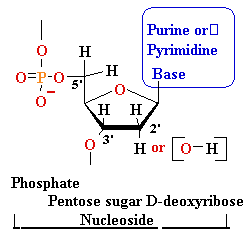



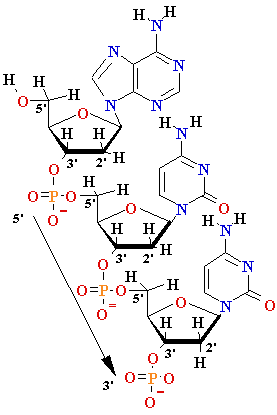
| nucleo-base |
nucleoside |
mono-nucleotide |
||||
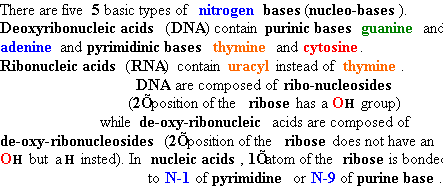
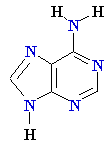
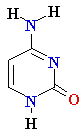
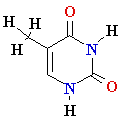
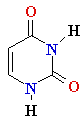
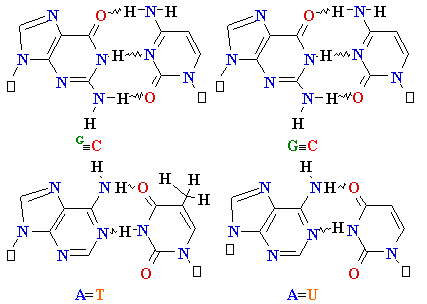
| Adenine (A) Guanine (G) Thymine (T) Cytosine (C) Uracyl (U) |
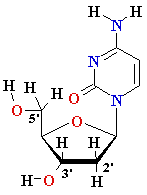
Deoxyadenosine
(dA) Deoxyguanosine (dG) Deoxythymidine (dT) Deoxycytidine (dC) Uridine (U) |
Deoxyadenosine
5’-phosphate (5’-dAMP) Deoxyguanosine 5’-phosphate (5’-dGMP) Deoxythymidine 5’-phosphate (5’-dTMP) Deoxycytidine 5’-phosphate (5’-dCMP) Uridine 5’-phosphate (5’- UMP) |
||||
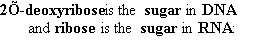
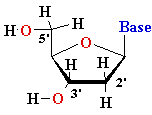
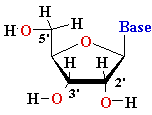
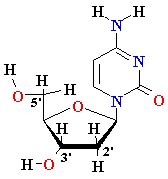


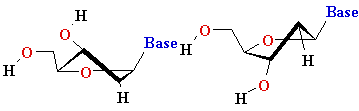
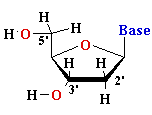

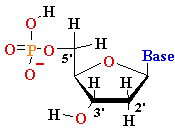

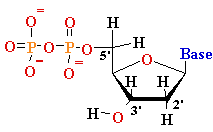
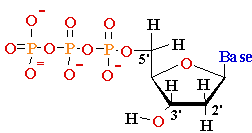


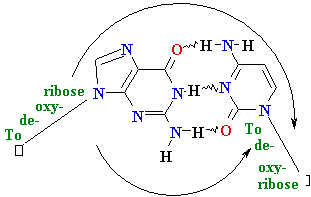

| Major
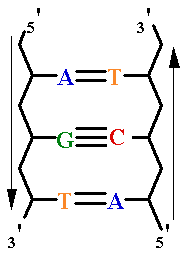
Groove of B-DNA • 12 Å wide, 8.5 Å deep • AT-nho • TA-ohn • GC-nho • CG-ohn |
Minor
Groove of B-DNA • 6Å wide, 7.5Å deep • AT-nho • TA-ohn • GC-nho • CG-ohn |
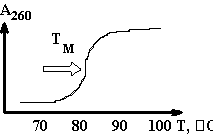

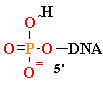
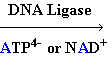
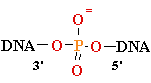 + AMP2- + H2O +
+ AMP2- + H2O +