Biochemistry of Medicinals I Phar 6151 CHAPTER TWO
Instructor: Dr. Natalia Tretyakova, Ph.D.
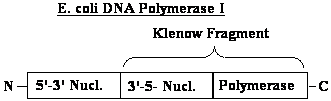
| Characteristic |
Pol I |
Pol II |
Pol III |
| Mol. Weight (Da) | 103 000 | 88 000 | 900 000 |
| Number of polypeptides | 1 | 4 | 10 |
| rate (nucleotides / sec) | 16-20 | 7 | 250-1000 |
| 3’--->5’ exonuclease | yes | yes | yes |
| 5’--->3’ exonuclease | yes | no | no |
| function | Primer removal, gap filling |
unknown | Major replicative polymerase |