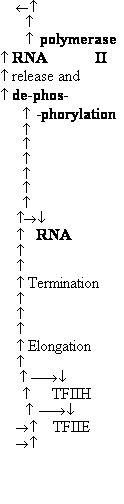| DNA |
RNA |
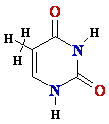 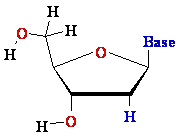 2'-de-oxy-ribose 2'-de-oxy-ribose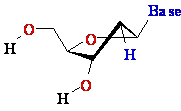 |
 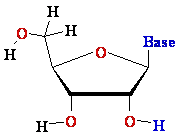 D-ribose D-ribose 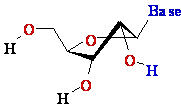 |
| Type |
Relative amount, % |
Sed.
Coeff. Sv |
Mass,
kD |
#
of Nucleotides |
| rRNA |
80 |
23 |
1.2•103 |
3700 |
| |
|
16 |
0.6•103 |
1700 |
| |
|
5 |
3.6•101 |
120 |
| tRNA |
15 |
4 |
2.5•101 |
75 |
| mRNA |
5 |
|
heterogeneous |
|
| Subunit |
Number |
Mass kD |
Function |
| a | 2 | 37 | Binds regulatory sequences |
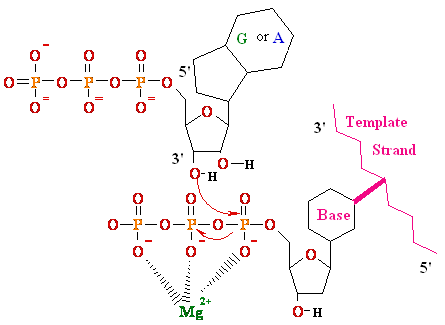
| b | 1 | 151 | Bonds NTPs, forms phosphodiester bonds |
| b' | 1 | 155 | Binds DNA template |
| holoenzyme | 1 | 70 | Recognizes promoter and initiates synthesis |

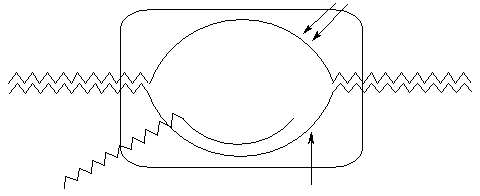
| Type |
Localization |
? RNA produced |
Inhibition
by amanitin |
| I |
Nucleolus | rRNA | Insensitive |
| II |
Nucleoplasm | mRNA | Strongly inhibits |
| III |
Nucleoplasm | tRNA,rRNA | Weakly inhibits |
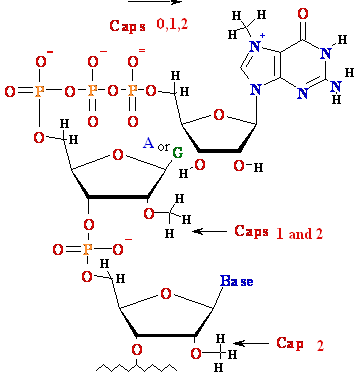

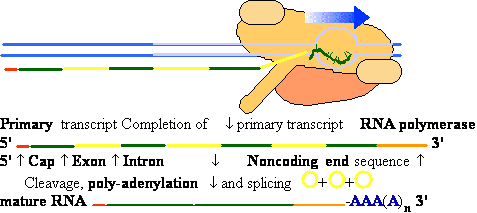
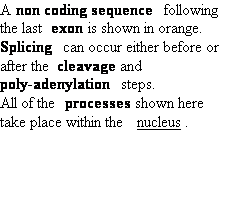
| |
Number of |
|
|
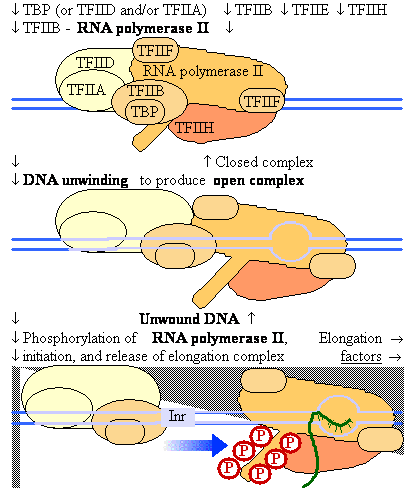
| Transcription factor |
subunits |
Subunit MW |
Functions |
| Initiation | |
|
|
| RNA polymerase II | 12 | 10 000-220 000 | Catalyzes RNA synthesis |
| TBP (TATA-binding | protein) 1 | 38 000 | Specifically recognizes the |
| TFIIA | 3 | 12 000,19 000, 35 000 |
Stabilizes binding of TFIIB and TBP to the promoter |
| TFIIB | 1 | 35 000 | Binds to TBP; recruits RNA polymerase-TFIIF complex |
| TFIID | 12 | 15 000-250 000 | Interacts with positive and negative regulatory proteins |
| TFIIE | 2 | 34 000, 57 000 | Recruits TFIIH; ATPase and helicase activities |
| TFIIF | 2 | 30 000, 74 000 | Binds tightly to RNA polymerase II; binds to TFIIB and
prevents binding of RNA polymerase to nonspecific DNA sequences |
| TFIIH | 12 | 35 000-89 000 | Unwinds DNA at promoter; phosphorylates RNA polymerase; recruits nucleotide-excision repair complex |
| Elongation * | |
|
|
| ELL‡ | 1 | 80 000 | |
| P-TEFb | 2 | 43 000,124 000 | |
| SII (TFIIS) | 1 | 38 000 | |
| Elongin (SIII) | 3 | 15 000, 18 000, 110 000 |
|