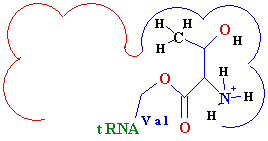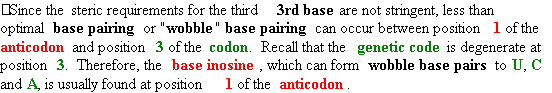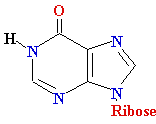
| |
U | C |
A |
G |
| U | UUU-Phe UUC-Phe UUA-Leu UUG-Leu |
UCU-Ser UCC-Ser UCA-Ser UCG-Ser |
UAU-Tyr UAC-Tyr UAA-Term UAG-Term |
UGU-Cys UGC-Cys UGA-Term-SelenoCys-Trp-Mitochondria UGG-Trp |
| C |
CUU-Leu CUC-Leu CUA-Leu CUG-Leu |
CCU-Pro CCC-Pro CCA-Pro CCG-Pro |
CAU-His
CAC-His CAA-Gln CAG-Gln |
CGU-Arg
CGC-Arg CGA-Arg CGG-Arg |
| A |
AUU-Ile AUC-Ile AUA-Ile AUG-Met |
ACU-Thr
ACC-Thr ACA-Thr ACG-Thr |
AAU-Asn AAC-Asn AAA-Lys AAG-Lys |
AGU-Ser
AGC-Ser AGA-Arg AGG-Arg |
| G |
GU-Val GUC-Val GUA-Val GUG-Val |
GCU-Ala
GCC-Ala GCA-Ala GCG-Ala |
GAU-Asp
GAC-Asp GAA-Glu GAG-Glu |
GGU-Gly GGC-Gly GGA-Gly GGG-Gly |
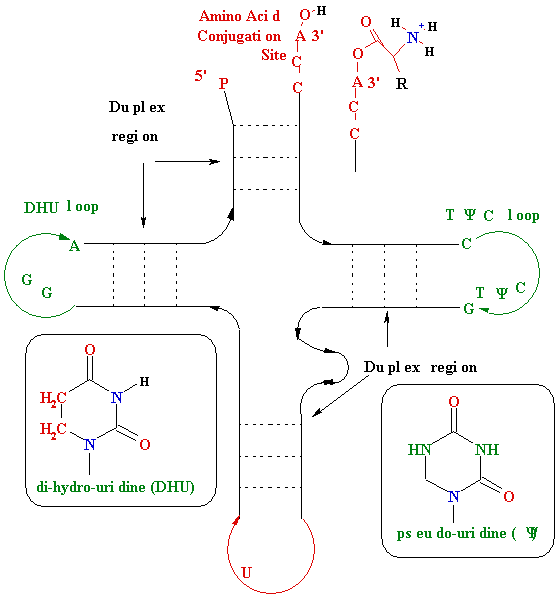
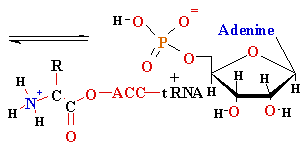
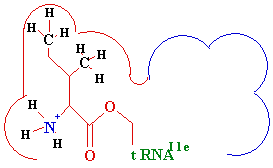 <--
Difference -->
<--
Difference -->
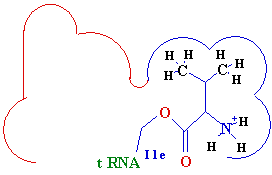
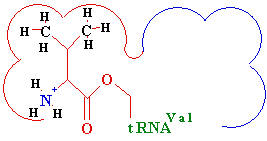 <---
Difference -->
<---
Difference -->