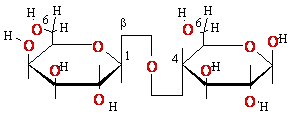
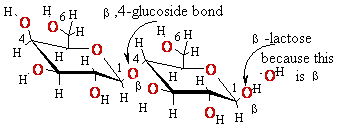
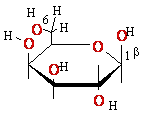
 +
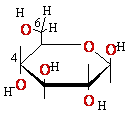
+ 
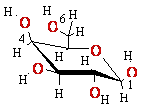 +
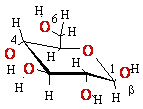
+ 
| P |
Promoter | Binding site for RNA Pol |
| O |
Operator | DNA control region that prevents transcription when repressor is bound |
| I |
Regulator gene | Codes for the repressor protein ◊ |
| z, y, and a | Structural genes | Code for b-galactosidase,
galactoside permease, and thio-galactoside trans-acetylase, respectively |
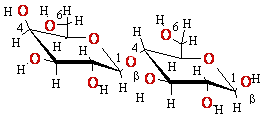
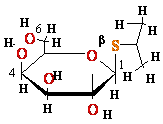
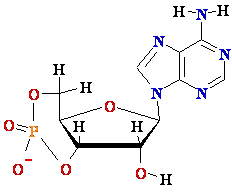 cAMP-
cyclic Adenosine Mono-Phosphate
cAMP-
cyclic Adenosine Mono-Phosphate