9.1 Peptide bond – primary structure
Proteins in cells synthesize ribosomes. Ribosomes are biocatalysts – combined enzymes complexes – biological sewing-machine of amino acids, which in polycondensation reaction with correct sequence of gene encoded one chain by peptide bonds bind each second fourteen of 20 different amino acids, forming dipeptides, tripeptides and polypeptides.
All 26000 encoded proteins synthesizes in ribosomes and first amino acid from messenger RNA molecule read methionine (Met[M]), with which start polycondensation reaction of polypeptide chain synthesis according on messenger RNA molecule encoded amino acid sequence. For example, linking to first amino acid methionine alanin produces dipeptide methioninalanin or in three letter abbreviation Met-Ala or in one letter symbol MA dipeptide and water:
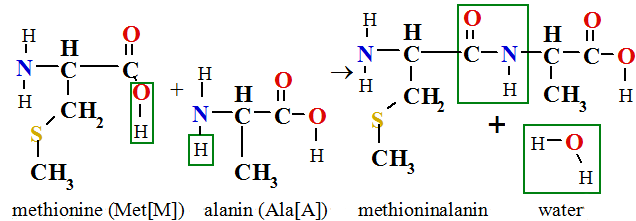
In polycondensation reaction arises water molecule, therefore the hydrolyze is reverse reaction. Hydrolyze reaction is reaction with water,
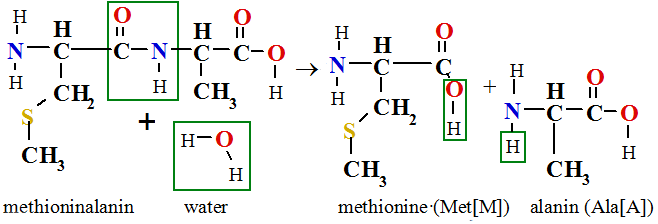
in which the hydrolyze products of polypeptide-protein are free amino acids solution in water. Therefore cooking meet in soup hydrolyzes free amino acid solution in water, what calls about bouillon.
Tripeptide alaninserincystein forms in polycondensation of three amino acids alanin, serine and cystein:
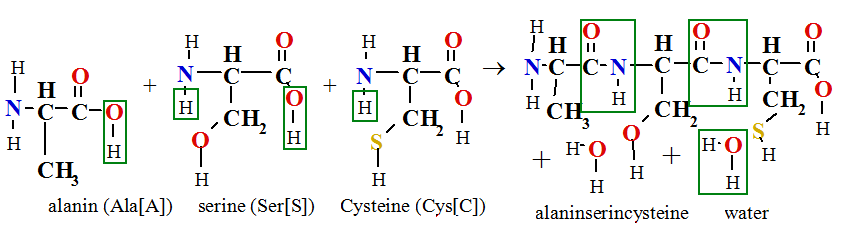
Three letters are Ala-Ser-Cys and one letter symbol abbreviation is ASC. Amino acid number one alanin on tripeptide chain has free amino group H2N- and its call about N terminal. C terminal is last amino acid cystein number three on chain with free carboxyl group -COOH.
9.2 Secondary, tertiary, quaternary structure of Proteins and five intermolecular forces
Five intermolecular forces strengthen folding of proteins in three different structure shapes, which calls one about secondary structure, tertiary structure and quaternary structure. In former chapter we consider primary structure of proteins, which forms polypeptide chains. Five intermolecular forces are:
1. and b) Hydrogen bonds,
2. and a) Salt bridge,
3. and d) Disulphide bonds,
4. and c) Hydrophobic bonds and
5. and e) Coordinative bonds.
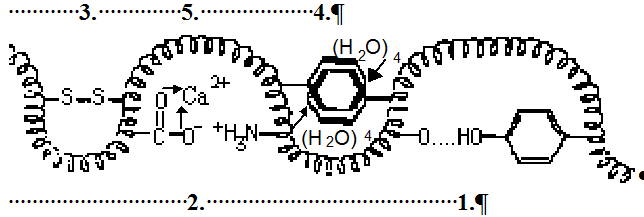
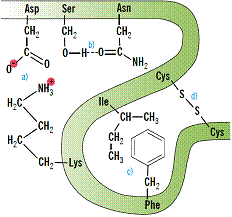
Fig.18 Stylistic picture of disulphide bond Cys S
S S
S Cys, coordinative donor acceptor bond calcium ion with carboxyl groups
Cys, coordinative donor acceptor bond calcium ion with carboxyl groups  COO→Ca2+←OOC
COO→Ca2+←OOC or iron(II) ion on center of hem →Fe2+←, salt bridge
or iron(II) ion on center of hem →Fe2+←, salt bridge
Asp COO----H3+N
COO----H3+N Lys, hydrophobic bond (H2O)4→00← (H2O)4 water squeeze together nonpolar residues 0 0 of amino acids, hydrogen bond Asn=O...H
Lys, hydrophobic bond (H2O)4→00← (H2O)4 water squeeze together nonpolar residues 0 0 of amino acids, hydrogen bond Asn=O...H O
O Ser.
Ser.
1. Hydrogen bond forms if between electronegative chemical elements oxygen atoms =O...H O
O or nitrogen atoms =N...H
or nitrogen atoms =N...H N= stand hydrogen atom, which covalently bind with one of atoms.
N= stand hydrogen atom, which covalently bind with one of atoms.
Oxygen or nitrogen atoms are hydrogen atom acceptors
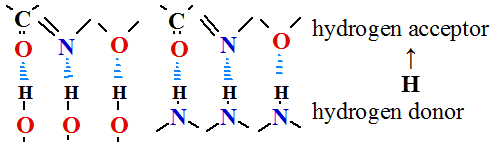
Oxygen or nitrogen atoms are hydrogen atom donors.
Secondary structure 2°
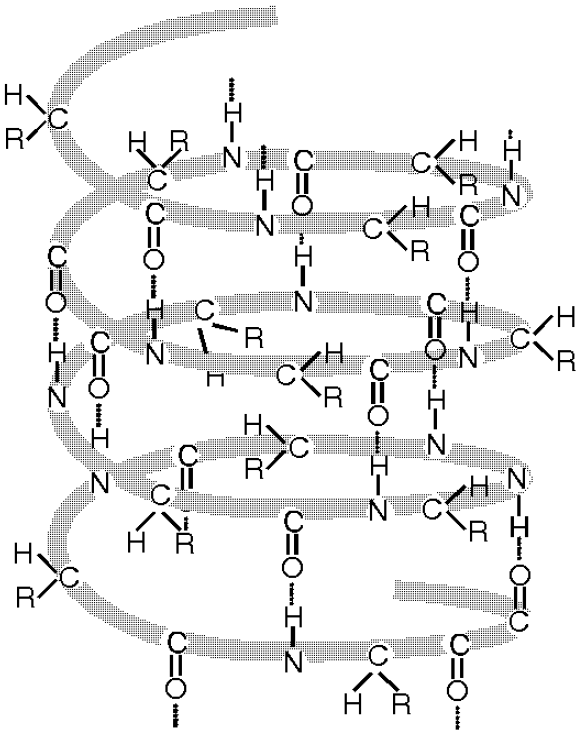
Hydrogen bond fastens secondary structure of alpha helixes, beta sheets and beta loops for proteins.
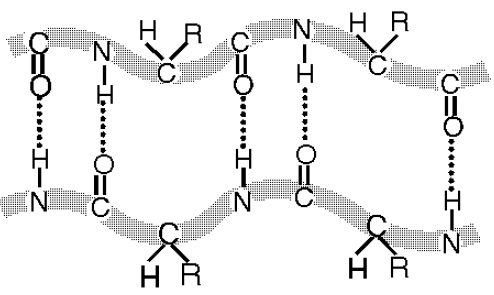 b
b
19 Fig. Polypeptide chain secondary structure of proteins folds together with hydrogen bonds into the alpha helix (a) and into the beta sheet (b).
19
2. Salt bridge forms between negative charged carbonic acid and positive charged neighbor on protein chains ammonium functional groups COO----H3+N
COO----H3+N
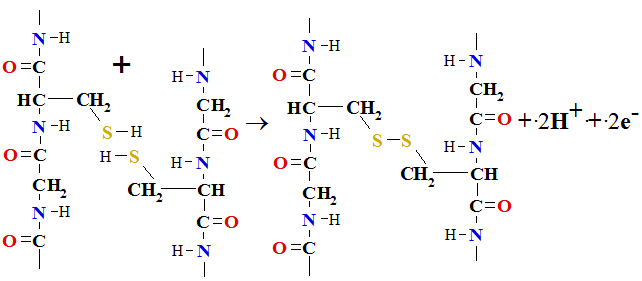
3. Disulfide bond forms under mild oxidation conditions between two protein chains joint opposite stand cystein (Cys[C]) amino acid oxidizing sulfhydryl groups
Cys SH +HS
SH +HS Cys => Cys
Cys => Cys S
S S
S Cys +2H+ + 2e-
Cys +2H+ + 2e-
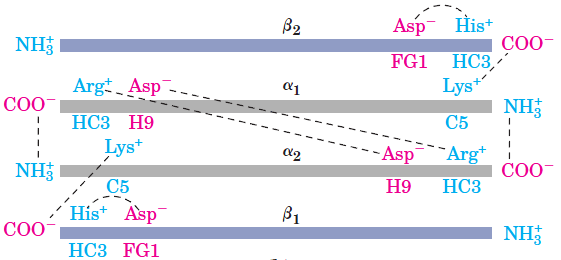
21 Fig. Venous blood hemoglobin has ten salt bridges, which joint four alpha1, alpha2, beta1 and beta2 protein chains with opposite charged negative carbonic acid  COO- and positive charged ammonium H3+N
COO- and positive charged ammonium H3+N functional groups.
functional groups.
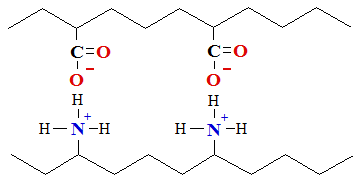
4. Hydrophobic bond forms in water medium. Meeting two protein chains and touching residues of nonpolar amino acids, for example, phenylalanine and leucine or isoleucine, water molecules press together with force, which is ten times stronger as Van der Walls forces. Hydrophobic force influences cooling of heated gelatin water solution, which forms jelly, similar as cooked legs or hade of pig in soup, which after cooling turns into jelly or (zilc in Latvian), because water press together nonpolar amino acids under influence of hydrophobic forces,
which lies in adjacent chains of neighboring mutual contacting proteins (polypeptide).
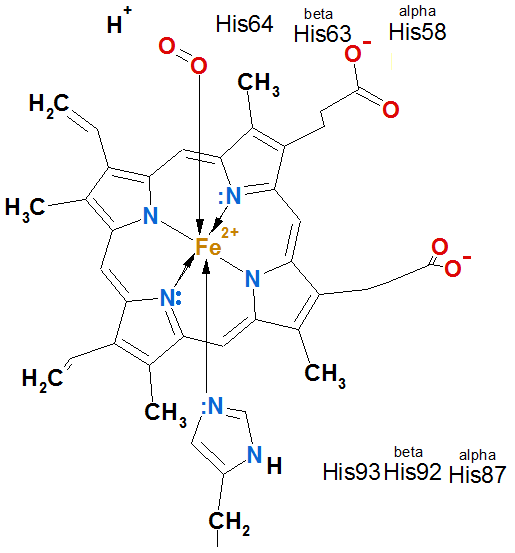
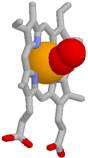
5. Coordinative bond form complex makers (look A.Rauhvarger, General Chemistry, vol.III, Complex compounds, part 12, p. 236) which are metallic ions: iron(II) ions Fe2+, iron(III) ions Fe3+, calcium ions Ca2+, magnesium ions Mg2+ also zinc ions Zn2+ or cooper ions Cu2+ and others, which are acceptors of donor oxygen and nitrogen unshared electron pairs, and, which (Fe2+,Fe3+, Ca2+, Mg2+) with coordination
number 6 or (Zn2+, Cu2+) with coordination number 4 coordinates around metallic ion 6 or 4 oxygen O and nitrogen N atoms from enveloping proteins, stabilizing tertiary and quaternary structure of proteins.
20
Tertiary structure 3° In tertiary structure folds secondary structure elements: alpha helixes, which resembles to tube of coiled protein chain, as well as beta sheets and beta loops, which provides parallel location tightly binding with hydrogen bonds of protein chains into beta sheets. In formation of tertiary structure take a place intermolecular interaction forces and some times all five: 1. Hydrogen bonds, 2. Salt bridges, 3. Disulfide bonds,
4. Hydrophobic bonds and 5.Coordinative bonds.
Quaternary structure 4° Quaternary structure is several protein separated chains aggregates, which bind together five intermolecular forces 1. Hydrogen bonds, 2. Salt bridges,
3. Disulfide bonds, 4. Hydrophobic bonds and
5.Coordinative bonds. For example:
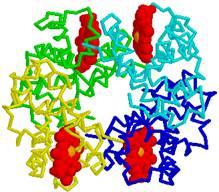
In hemoglobin molecule four protein chains of tertiary structure alpha1, alpha2, beta1 and beta2 binds 1. Hydrogen bonds, 2. Salt bridges, 4. Hydrophobic bonds and 5.Coordinative bondsFe2+.
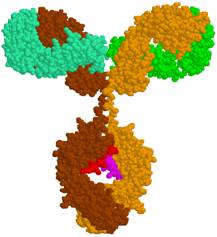
In immunoglobulin molecule two heavy and two light protein chains of tertiary structure bind :1. Hydrogen bonds,
2. Salt bridges, 3. Disulfide bonds, 4. Hydrophobic bonds.
Denaturation of proteins
Destruction of protein quaternary, tertiary and secondary structure is called as well denaturation. For example, egg white is transparent fluid viscous liquid, which natural appearance determines included proteins primary, secondary, tertiary and quaternary structure. Boiled eggs white are white congealed mass, because high temperature boiling breaks four intermolecular forces: 1. Hydrogen bonds, 2. Salt bridges, 4. Hydrophobic bonds and 5.Coordinative bonds.
3. Disulfide bonds demolish only at presence of reducing agents and present disulfide bonds in curdle can not break with heating only. Therefore heating curdle can obtain next cooking product cheese.
To prepare meal humans have learned denaturate proteins for nutrition, which perfect would be used in food containing amino acids, because organism absorbs just free amino acids. Therefore peoples in cooking meal apply the same methods as in chemistry labor methods: separation or grind into smaller peaces, heating and boiling, adding of acids, for example, acetic acid, citric acid or vine addition, in which always are present acids.
Mentioned denaturation actions with food applied proteins demolish quaternary, tertiary and secondary protein structure, but reaction of hydrolyze break the primary structure and release free amino acids, which absorb human organism from food prepared meals, that inside cells in ribosomes as new would synthesize for organism necessary proteins.
21






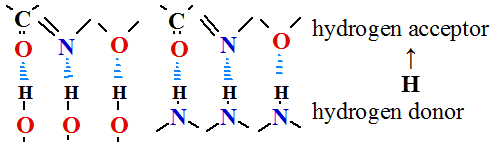
 b
b

 amino acid
amino acid