| Preformed nucleotides can be recycled by salvage pathways
|
| FOR MANY ORGANISMS SALVAGE PATHWAYS ARE THE PRINCIPAL SOURCE OF NUCLEOTIDES |
| The observation that organisms blocked in the de novo biosynthetic pathways can survive and grow if a source of nucleotides is available from the diet indicates the strategic importance of the nucleotide-salvage pathways. The salvage pathways are especially important for many parasites. These organisms prey metabolically on their host, using preformed metabolites, including nucleotides. Some parasites, such as Mycoplasma, Borrelia, and Chlamydia, have lost the genes required for the de novo synthesis of nucleotides; they obtain these important components from their host. |
| Even in humans, resting T lymphocytes, immune-system cells produced in the thymus, meet their metabolic requirements for nucleotides through the salvage pathway, but de novo synthesis is required for lymphocyte proliferation. The salvage of nucleotides is especially important in HIV-infected T lymphocytes. In asymptomatic patients, resting lymphocytes show a block in de novo pyrimidine biosynthesis, and correspondingly reduced pyrimidine pool sizes. Following activation of the T-lymphocyte population, these cells cannot synthesize sufficient new DNA. The activation process leads to cell death, contributing to the decline in the T-lymphocyte population during the late stages of HIV infection. |
In addition to de novo synthesis, cells can use preformed nucleotides obtained from the diet or from the breakdown of endogenous nucleic acids through salvage pathways. In
mammals, there are two enzymes in the purine salvage pathway. Adenine phosphoribosyltransferase (APRT) converts free adenine into AMP:
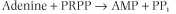
|
Hypoxanthineguanine phosphoribosyltransferase (HG-PRT) catalyzes a similar reaction for both hypoxanthine and guanine:
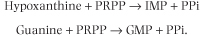
|
| Purine nucleotides are synthesized preferentially by salvage pathways, so long as hypoxanthine is available. This preference is mediated by hypoxanthine inhibition of amidophosphoribosyl transferase, step 2 in the de novo pathway (Fig. 29.2).
|
|