| Denaturant-gradient gel electrophoresis (DGGE)
|
| DGGE uses 'melting' of double-stranded DNA
|
|
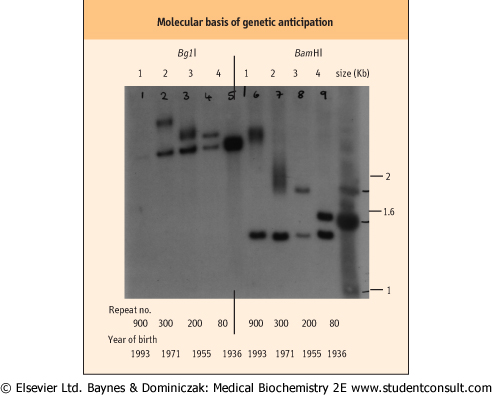
| Figure 34.17 Molecular basis of genetic anticipation. Myotonic dystrophy (MD) exhibits anticipation within families, i.e. the clinical features of the disease become progressively more severe and its age at onset of symptoms is younger with each successive generation. The underlying problem is the presence of an unstable trinucleotide expansion within the MD gene, which progressively expands with each meiosis. This results in the inheritance of larger mutations, which give rise to a more severe clinical picture with the tendency for the features to manifest at an earlier stage. The Southern blot illustrates the progressive increase in size of the trinucleotide expansion in successive generations of a family with MD. The normal allele size is 3.4 kb for the BglI digest and 1.4 kb for the BamHI digest. It is clear, using both restriction enzymes, that with each generation there is a progressive expansion in the size of the trinucleotide repeat. (This is a good example of an RFLP.) |
In SSCP, changes in the tertiary structure of single-stranded DNA can result in differing electrophoretic mobilities. However, this type of analysis cannot be used for double-stranded DNA. Nevertheless, if double-stranded DNA undergoes
electrophoresis in a gel that contains a gradient of increasing amounts of denaturant, e.g. urea , double-stranded DNA will then partially denature, to form single-stranded regions and so undergo a marked change in mobility. A single nucleotide change between two alleles of a gene will affect the melting point of the alleles and thus the mobilities of amplified PCR fragments in this kind of denaturing gel. , double-stranded DNA will then partially denature, to form single-stranded regions and so undergo a marked change in mobility. A single nucleotide change between two alleles of a gene will affect the melting point of the alleles and thus the mobilities of amplified PCR fragments in this kind of denaturing gel.
|
| page 491 |  | | page 492 |
| PROTEIN TRUNCATION TESTING IN BREAST CANCER |
| A hereditary form of breast cancer associated with the coexistence of ovarian cancer is caused by mutations in the BRCA1 gene. This gene, a tumor suppressor gene, was discovered following the analysis of large pedigrees with breast/ovarian cancer. Analysis of the mutations found in the genes of affected individuals showed that most of the mutations were either nonsense mutations, altered splice sites, or frameshift mutations, which gave rise to abnormally truncated protein products. Following identification of the common mutation types in BRCA1 (and the subsequently discovered BRCA2), a protein truncation test has become a standard procedure for examining both pathologic tissue and blood from individuals at risk for the mutation. |
| Comment. The translation of mRNA to the final protein product results in a peptide of predictable size. If a gene is disrupted or has an insertion, deletion or a frameshift mutation that produces a premature stop codon, which occurs with 1/64 frequency in a random polynucleotide sequence, then the resulting peptide will be abnormally small. This difference can then be detected by electrophoresis of the protein product in a suitable gel and demonstrating the presence of a truncated protein product. |
| To perform DGGE, PCR is performed using radiolabeled primers that have a GC clamp at their ends. This is a tail of guanine and cytosine residues added to the PCR primers such that the PCR product will have ends that have artificially created GC ends. These GC ends, or clamps, improve the sensitivity by preventing complete melting of double-stranded
DNA. PCR products are electrophoresized in the denaturing gel and, following autoradiography, different alleles of the suspect gene are identified (Fig. 34.18). This is particularly useful for rapid scanning of individually amplified exon units from a large gene with many introns.
|
|