| Primary structure of proteins
|
| The primary structure of the protein is the linear sequence of amino acids
|
| Glutathione (GSH) is a tripeptide with the sequence l-γ-glutamyl-l-cysteinylglycine (Fig. 2.6). If the thiol group of the cysteine is oxidized, the disulfide GSSG is formed. GSH is the major peptide present in the cell. In the liver, the concentration of GSH is ∼5 mmol/L. GSH plays a major role in the maintenance of proteins in their reduced forms and in antioxidant defenses (Chapter 35). The enzyme γ-glutamyl transpeptidase is involved in the metabolism of glutathione and is a marker for some liver diseases, including hepatocellular carcinoma and alcoholic liver disease. |
 |
|
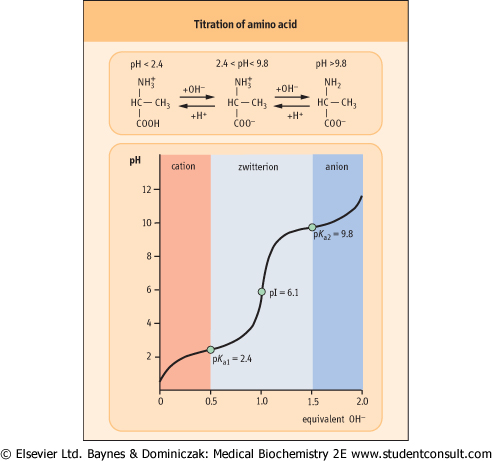
Figure 2.4 Titration of amino acid. The curve shows the number of equivalents of NaOH consumed by alanine while titrating the solution from pH 0 to pH 12. Alanine contains two ionizable groups: an α-carboxyl group and an α-amino group. As NaOH is added, these two groups are titrated. The pKa of the α-COOH group is 2.4, whereas that of the α-NH3+ group is 9.8. At very low pH, the predominant ion species of alanine is the fully protonated form:
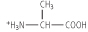 At the mid-point in the first stage of the titration (pH 2.4), equimolar concentrations of proton-donor and proton-acceptor species are present, providing good buffering power.
At the mid-point in the first stage of the titration (pH 2.4), equimolar concentrations of proton-donor and proton-acceptor species are present, providing good buffering power.
 The second stage of the titration corresponds to the removal of a proton from the -NH3+ group of alanine. The pH at the mid-point of this stage is 9.8, equal to the pKa for the -NH3+ group. The titration is complete at a pH of about 12, at which point the predominant form of alanine is:
The second stage of the titration corresponds to the removal of a proton from the -NH3+ group of alanine. The pH at the mid-point of this stage is 9.8, equal to the pKa for the -NH3+ group. The titration is complete at a pH of about 12, at which point the predominant form of alanine is:
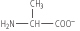 The pH at which a molecule has no net charge is known as its isoelectric point, pI. For alanine, it is calculated as:
The pH at which a molecule has no net charge is known as its isoelectric point, pI. For alanine, it is calculated as:
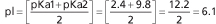 |
| page 12 |  | | page 13 |
| Figure 2.5 Structure of a peptide bond. |
| Figure 2.6 Structure of glutathione. |
In proteins, the primary amino group of one amino acid is linked to the carboxyl group of the next amino acid, forming
an amide (peptide) bond (Fig. 2.4). During the formation of a peptide bond, a molecule of water is eliminated, as shown in Figure 2.5. The amino acid units on a peptide chain are referred to as amino acid residues. A peptide chain consisting of three amino acid residues is called a tripeptide, e.g. glutathione in Figure 2.6. By convention, the amino terminus (N-terminus) is taken as the first residue, and the sequence of amino acids is written from left to right. When writing the peptide sequence, one uses either the three-letter or the one-letter abbreviations of amino acids is written from left to right. When writing the peptide sequence, one uses either the three-letter or the one-letter abbreviations of amino acids , such as Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu or D-R-V-Y-I-H-P-F-H-L (see Table 2.1). This peptide is angiotensin, a peptide hormone that affects blood pressure. The amino acid residue having a free amino group at one end of the peptide, Asp, is called the N-terminal amino acid (amino terminus), whereas the residue having a free carboxyl group at the other end, Leu, is called the C-terminal amino acid (carboxyl terminus). Proteins contain between 50 and 2000 amino acid residues. The mean molecular mass of an amino acid residue is about 110 dalton units (Da). Therefore the molecular mass of most proteins is between 5500 and 220 000 Da. Human carbonic anhydrase I, an enzyme that plays a major role in acid-base balance in blood (see Chapter 23), is a protein with a molecular mass of 29 000 Da (29 kDa). , such as Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu or D-R-V-Y-I-H-P-F-H-L (see Table 2.1). This peptide is angiotensin, a peptide hormone that affects blood pressure. The amino acid residue having a free amino group at one end of the peptide, Asp, is called the N-terminal amino acid (amino terminus), whereas the residue having a free carboxyl group at the other end, Leu, is called the C-terminal amino acid (carboxyl terminus). Proteins contain between 50 and 2000 amino acid residues. The mean molecular mass of an amino acid residue is about 110 dalton units (Da). Therefore the molecular mass of most proteins is between 5500 and 220 000 Da. Human carbonic anhydrase I, an enzyme that plays a major role in acid-base balance in blood (see Chapter 23), is a protein with a molecular mass of 29 000 Da (29 kDa).
|
|