I. Introduction 1J4N,4KSC with 10 water molecules
H1,H2,HB,H3,H4,H5,HE,H6,H7,A,B,C,D,E,LB,LE
Human abundant water channel aquaporin-1 (AQP1). Backbone thin
Primary 1° structure of monomere polypeptide chain 1YMG is
constitute of 233 amino acids on the linear chain
by 232 peptide bonds Primary p.12 on backbone chain of 233 alpha Carbons of amino acids
Polypeptide chain is folded
into six+ two HB,HE half trans membrane units
called as helicies H1,H2,H3,H4,H5,H6,HB,HE forms
secondary 2° structure using Hydrogen bonds. Secondary p.13
The folding secondary structure 9 helicies using Hydrogen,
Hydrophobic bonds and some Salt Bridges forms Tertiary 3° and Quaternary 4°
structer one monomer subunit for quaternary tetramere 2B5F.
Short helix 9th H7in large loop C
Tertiary-Quaternary p.14-p.15
Tetramere Quaterbary structure 4° of molecule
2B5F from Protein Data Bank is formed from
four monomere molecules like 1YMG deponed and taken
from Protein Data Bank as four subunits in AQP0 quaternary 4̊
structure composition assambled biological membrane protein for
water, O2, NO, CO small molecules transport across the
biological membranesTertiary-Quaternary p.14-p.15
Aquaporins are large families (over 450 members) that
are present in all kingdoms of life. Protein folding
supporting intermolecular forces are five :
1. and b) Hydrogen bonds,
2. and a) Salt bridge,
3. and d) Disulfide bonds,
4. and c) Hydrophobic bonds and
5. and e) Coordinative bonds.
Bos taurus AQP0 remains a tetramer.
Its 4-fold axis in the crystal is aligned with the c axis, and
thus, the plane of the membrane would lie perpendicular
to the c axis. In the plane of the membrane, the tetramer is
˜60 Å wide (˜74 Å, corner to corner) and ˜53 Å tall.
Each b AQP0 monomer is ˜35 Å in diameter sise and
contains one channel at its center that is oriented parallel
to the 4-fold axis of the tetramer from Protein Data Bank, as
in other AQPs 2B5FMarz SPINACH plant Aquaporin.
H1,H2,HB,H3,H4,H5,HE,H6
Backbone thin
1J4N,4KSC with 10 water molecules
Human AQP1 1J4Nbio1. 1J4Nbio1Marz. remains a tetramer.
Its 4-fold axis in the crystal is aligned with the c axis, and
thus, the plane of the membrane would lie perpendicular
to the c axis. In the plane of the membrane, the tetramer is
55.7 to 57 Å wide ( 69.1 to 72.7Å, corner to corner) and penetrate
the membrane through its tikness
51.6 Å tall.
Each AQP1 monomer 1J4N is 25.5 Å in centre diameter as
Each AQP1 monomer 4CSK is 25.5 Å in centre diameter as
well vestibule diameter
25.5 Å to 36.8 Å and
contains one channel at its center that is oriented parallel
to the 4-fold axis of the tetramer, as in other AQPs .
Primary 1° structure AQP1 of 269 monomeres on polypeptide chain
1J4N present
constitute of 1-249 amino acids on the linear chain
by 248 peptide bonds
4KCSK present
constitute of 3-235 amino acids on the linear chain
by 232 peptide bonds
Primary p.12
Polypeptide chain is folded
into six+ two half trans membrane units
using Hydrogen bonds forms secondary 2° structures called as helicies
H1,H2,HB,H3,H4,H5,HE,H6,H7 forms. Secondary p.13
Six transmembrane domains (TMDs), highly hydrophobic, with alpha-helix
structure and five connecting loops. The alpha-helices are named from
the N-end succesively H1,H2,H3,H4,H5
C-terminal helix H6, H7 and the
five loops are named A,B,C,D,E
The NPA boxes are located in the loops LB and LE,
which are rather hydrophobic in nature and have short
(half) helices HB and HE
The six TMDs (tilted at about 30° with respect to the
membrane normal right-handed) form a right-handed bundle enclosing
the channel (pore) formed by the NPA motifs and the
short tetramer helices HB and HE, bent into the
six-helix bundle and connected in the center of the bilayer.
4 water molecules
Backbone thin
The TMDs and the loops form a core (embedded in the
membrane lipid bilayer), to which two “legs” (represented
by the cytosolic N- and C-ends) are attached.
The folding secondary structure 9 helicies using Hydrogen,
Hydrophobic bonds and some Salt Bridges forms Tertiary 3°
structer one monomer subunit for quaternary tetramere 1J4Nbiol.
Tertiary-Quaternary p.14-p.15
Tetramere molecule
1J4Nbiol from Protein Data Bank is formed
from
four monomere molecules 1J4N deponed and taken
from Protein Data Bank as four subunits in AQP1 quaternary 4̊
structure composition assambled biological membrane protein for
water,
O2, NO, CO small molecules transport across the
biological membranes Tertiary-Quaternary p.14-p.15
1J4Nbio1Marz. Aquaporins are large families (over 450 members) that are present in all kingdoms of life.
H1,H2,HB,H3,H4,H5,HE,H6,H7,A,B,C,D,E,LB,LE
Protein folding supporting intermolecular forces are five :
1. and b) Hydrogen bonds,
2. and a) Salt bridge,
3. and d) Disulfide bonds,
4. and c) Hydrophobic bonds and
5. and e) Coordinative bonds.
II. Structure of 263 amono acids 1J4NMarz
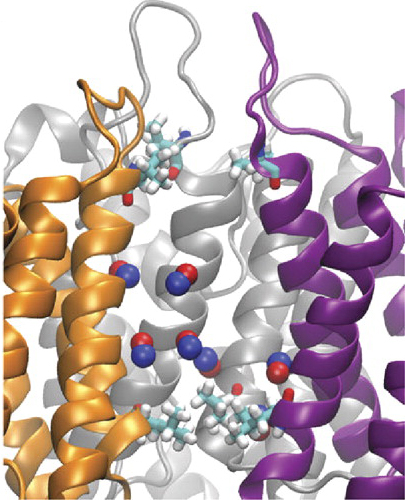
NPA motifs and the short tetramer helices HB:78-88 and HE:194-205,
HB,HE,LB,LE
bended into the six-helix bundle and connected in the center of the bilayer.
H1,H2,H3,H4,H5,H6,
Cterminal helix H10,H11
five loops are named
A,B,C,D,E,
The NPA boxes are located in the loops LB:206-211 and LE:191-193,
which are rather hydrophobic in nature and have short
(half) helices HB and HE
The six trans-membrane domaines TMDs Backbone thin
(tilted at about 30° with respect to the membrane normal) form a
right-handed bundle enclosing the channel (pore) formed by the
NPA boxes (or motifs) with three amino acid residues The regions containing the
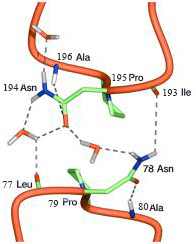
Two NPA motifs are LB-HB and LE-HE, respectively. They are highlighted by darker color.
The positions of the Ca atoms of Asn78,Pro79,Ala80 and Asn194,Pro195,Ala196 are indicated by spheres.
(asparagine, proline, alanine: Asn-Pro-Ala)
LE-HE & LB-HB short (half) helices HB and HE
Asn194,Pro195,Ala196-Ala75,Asn78,Pro79,Ala80
The two asparagines at the positive ends of helices HB and HE act as
hydrogen donors for hydrogen bonding to the oxygen atom of the
Water molecule in the center of the pore on
The mechanism of water selectivity and proton discrimination remains highly
controversial. Possible factors that have been proposed to contribute to the
selectivity include the small size of the constriction, the arrangement of
helical dipole moment near the constriction, and the energetic cost of
bringing positively charged H3O+ ions into the cell membrane.
The breakage of hydrogen bonds in the water flow, caused either by the
competing interactions of the conserved residues Asn78 and Asn194, or
caused by the curvilinear nature of the aqueous pathway (4), also has been suggested
to prohibit the formation of the single-file water network that would be required for efficient proton translocation.
The Water molecule is oriented perpendicular to the pore axis;
the central Water molecule forms (by its oxygen) hydrogen bonds with the
amido groups of Asn78 and Asn194 this Water molecule can only engage in
on
hydrogen bonding leading outwards from the center of the pore toward the
extracellular and the cytoplasmic entrance of the pore. The lines of Water
molecules in the two pore halves thus have opposite hydrogen bond polarity,
preventing protons to cross the central Water molecule.
Hydrophobic channel walls direct water flow into single file by 15 amino acids.
One said chains
Leu85(HB),Ile174(H5)
near-isosteric Leu hydrophobic Ile residues &
other said the hydrophobic lining of the H2O pathway includes
Phe24,Ile25,Ile29(H1),Phe58(H2),Ile62(H2),Ala75(LB),Leu77
Significant water density was observed in the space near H4 and H5 as
well.
This pathway also is predominantly lined with hydrophobic residues such as
Val81(HB),Leu151(H4),Val155(H4),Val178(H5),Ile172(H5), Ile193(LE)
1J4N H1,H2,HB,H3,H4,H5,HE,H6,H7,A,B,C,D,E,LB,LE
Six Water molecules form a single file through the pore.
on
The aqueous pathway are lined with hydrophobic residues. In a wider hydrophobic pathway, the cohesive forces among the polar water molecules are ber than the adhesive forces between the polar water molecules and the hydrophobic wall, so that the water flow tends to form a “tube” that enables water molecules to flow quickly in the axial direction of the pathway without becoming stuck to the surrounding wall. However, in the region of the narrow constriction of AQP1, the water flow is as thin as a single water molecule. It is then essential to have polar side chains to maintain the hydrogen-bonding interactions.
2-fold symmetry in that NPA region, roughly pointing to the
Two aqueous pathways, respectively. Pro79 and Pro195 on the NPA
motifs are completely symmetric on HB/HE contact
Pro79 and Pro195 in the NPA motifs may be important for maintaining the
sharply curved conformations of loops LB and LE
As shown, the Od atom of Asn78 side chain forms a hydrogen bond with
the main chain N–H group of Ala78, and the side chain Nd–H group of
Asn76 forms a hydrogen bond with the main chain carbonyl group C=O of Leu77.
Similarly, the Od of Asn194 side chain forms a hydrogen bond with the main chain
N–H group of Ala196, and the side chain Nd–H group of Asn194 forms a
hydrogen bond with the main chain carbonyl group C=O of Leu77
on
As the constriction opens up, water molecules can step in and form hydrogen bonds
with Asn78 and Asn194
1J4N H1,H2,HB,H3,H4,H5,HE,H6,H7,A,B,C,D,E,LB,LE
HB/HE Pro79 and Pro195 on
Phe24 and Phe58 seem to play an important role in directing the water flow near the NPA motifs.
Phe24, significantly influence the size of the constriction as Phe24 has significantly influence
to the size of the constriction. Only when the distance between the side
chains of Phe24 and
Asn194 is larger than 5.0 Å are there water molecules in the
constriction, hydrogen-bonded to the side chain and main chain of Asn194.
His182, which is located close to Phe58, has been found to be conserved
among all of the water-selective aquaporins, but is frequently replaced with a Gly in
glycerol-conducting channels.
His182 plays a particular role in water conduction. It was observed that the
side chain of His182 participates in hydrogen-bonding interactions with
water molecules in the constriction region and forms stable hydrogen bonds with the
main chain carbonyl groups of Val178(H5) and Gly192(LE). Here,Gly192
quickly flipped its backbone conformation from that in 1J4N within 50 ps.
So that its interaction with His182 became possible. Given the fact that
His182 and Asn194 are the only Two polar residues near the constriction region.
Asn78 and Asn194 are just right for the size. There is not enough space to
accommodate a larger side chain in the vicinity of the Calpha atoms of Asn78 and Asn194. The Gly192 C=O there would make the loops too floppy with His182
The physical limitation on the size of substrates allowed to permeate the AQP1 pore is
imposed by the 3 Å diameter of the narrowest region of the pore, which is only
slightly larger than the 2.8 Å diameter of the Water molecule. The pore
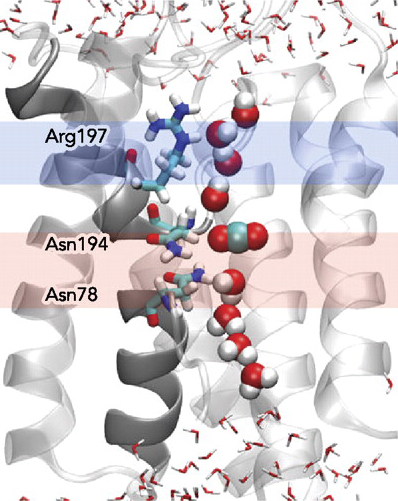
constriction Specifity Filter SF prevents permeation of all molecules bigger than Water,
including hydrated ions. The narrowest region of the pore in AQP1 was
named the Ar/R constriction site, because it contains highly conserved
aromatic and arginine residues. The Ar/R constriction site is formed in
hAQP1 by Arg197, His182, Phe58, and Cys191
Arg 197 and His182 line one side of the pore creating a hydrophilic
surface, whereas the Phe58 is located on the opposite side hydrophobic surface.
Extracellular Cys191 in the second NPA loop is the site of binding of
mercurial Hg2+ reagents that leads to reversible blockage of water transport.
Cys191 is the site for the inhibition by mercurials of Water permeation through
the pore. The mercurial-sensitive C191 and the analogous
A75 (colored red) are indicated
on
This structure is called the aquaporin fold.
So the channel (pore) is a narrow tunnel in the center of the
molecule, that has at the extracellular and cytoplasmic faces
funnel-shaped openings (atria telpa or vestibules priekštelpa).
The surface of the AQP1 pore is formed by highly
conserved residues in ,H2 and H5 and the C-terminal halves of the
H1 and H4 (forming the remaining surface of the pore).
This model was called “hourglass model”.
10 water molecules atoms of oxygen
301,302,303,304,346,361,381,393,397,414
on
Six Water molecules form a single file through the pore.
Ala196,Pro195,Ala197-Asn194,Asn78 -Arg197,His182,Phe58,His76-Cys191 on NPA,SF
301,302,303,304,346,361 on
The physical limitation on the size of substrates allowed to permeate the AQP1
pore is imposed by the 3 Å diameter of the narrowest region of the pore, which is only
slightly larger than the 2.8 Å diameter of the Water molecule.
The pore constriction prevents permeation of all molecules bigger than
Water, including hydrated ions. The narrowest region of the pore in
AQP1 was named the Ar/R constriction site, because it contains highly
conserved aromatic and arginine residues.
Water permeation through the pore.
Despite its extreme Water permeability, allowing permeation of
3 × 109 Water molecules per monomer per second, AQP1 (and
other WCPSs)
strictly prevents the conduction of protons. This is physiologically very important,
as the passage of protons through the pore would anihilate
the proton gradient across the cell membrane that serves as a major energy transfere mechanism from stored energy molecules fatty acids and glucose in addipose fatt tissues and liver glycogene.
The proton exclusion may be seen as the most exceptional feature of AQPs,
and the NPA motifs play an important role.
The two asparagines at the positive ends of helices HB and HE act as
hydrogen donors
to the oxygen atom of the Water molecule in the center of the pore.
The water molecule is oriented perpendicular to the pore axis;
the central water molecule forms (by its oxygen) hydrogen bonds
with the amido groups of Asn76 and Asn192;
this water molecule can only engage in hydrogen bonding leading
outwards from the center of the pore toward the extracellular and the
cytoplasmic entrance of the pore. The lines of water molecules in the
two pore halves thus have opposite hydrogen bond polarity, preventing
protons to cross the central water molecule.
The electrostatic proton
barrier in AQPs involves not only the NPA motifs, but also the Ar/R constriction size.
Mutation experiments showed that removal of the
positive charge from the Ar/R constriction site in two AQP1 mutants,
Arg197Val and His182Ala/Arg197Val, appeared to allow the passage
of protons through the AQP1 pore.
Arg197,His182
His71,His76,Arg162,Arg163,Arg164,Thr159
301,302,303,304,346,361 on
The positive charges of -NH2+ an arginine Arg197 residue and
at the histidine His182 of extracellular vestibule
as well as residues His71,His76,Arg162,Arg163,Arg164,Thr159
in the cytoplasmic vestibule would also help
to repel protons from entering the pore. In addition to these electrostatic
factors another major source of the barrier for proton transport in AQPs is
associated with the loss of the generalized solvation energy upon moving the
proton charge from the bulk solvent to the center of the channel.
H2O,
O2, NO, CO.
On the other hand the CO2 permeability of AQP1 is
controversial, even in recent publications, particularly in
regard with its physiological significance.
Transport of CO2 by some plant AQPs was reported.
Is conventional oxygen transport through WCPSs AQPs O2.
In addition, evidence for passage of NO through the AQP1
was published.
Other WCPSs are permeable for H2O2,
ammonia NH3,antimonite,arsenite,silicic acid, as well toxic CO.
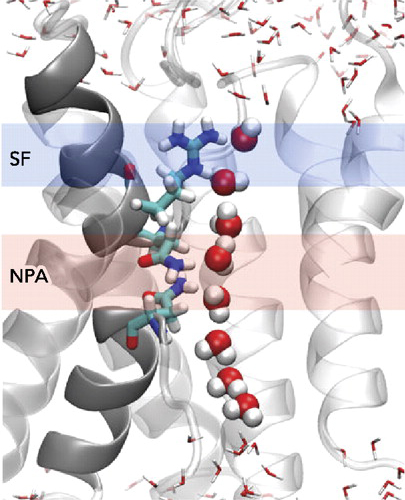
III. The 28-Å-long, cylindrical AQP1 channel is flanked by shallow vestibules
The 28-Å-long, cylindrical bAQP1 channel is flanked by shallowvestibules on each end. Channel volume is shown in the background,
with major channel-forming residues. The pink central region
has a diameter of <2.5 Å, the blue regions both side has a diameter of
>2.5 Å and <10 Å long distance from pink central region region cener 0.
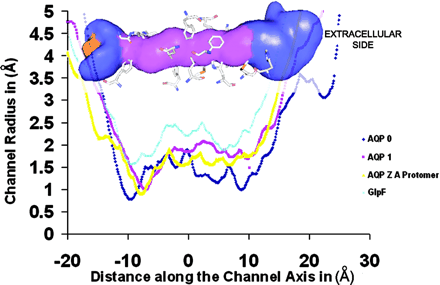
Starting from the extracellular side, the vestibule narrows to a diameter of <10 Å
1J4NMarz 4 water molecules
between residues
are oriented into the center line of the channel and is responsible for most
of the narrowing of the vestibule. The channel narrows a diameter of 1.99 Å .
This region is the narrowest region of the channel (Graph).
References
1. Int J Mol Sci.2018.Jun;19(6):1577.4CSK,4OJ2,1J4N
1. Acta Crystallogr F Struct Biol Commun.2014;70(Pt 12):1657-63 1J4N
1. Physiology June 1, 2010 vol. 25 no. 3 142-154 1J4N
2.Mol Biol Evol (2011) 28 (11): 3151-3169. Volume 28,, Issue 11 Pp. 3151-3169. 1J4N
3.Proc Natl Acad Sci U S A. 2006 January 10; 103(2): 269–274. Biochemistry 1J4N
4.Proc Natl Acad Sci U S A. 2001 December 4; 98(25): 14345–14349. 1FQY-1HW0,1J4N
Back to the Index...