| The principles of molecular hybridization
|
| In molecular hybridization, it is essential that the probe and target are initially single-stranded
|
| Probes can vary in both their size and their nature (Table 34.1). However, one essential feature of any hybridization reaction is that both the probe and the target must be free to base pair with one another. The process of separating the two strands of DNA is called DNA denaturation or melting. Probe DNA is generally denatured by heating. If the target DNA is double-stranded then it too must be denatured, either by heating or treatment with alkali. (NB: alkali is not used to denature RNA because it leads to hydrolysis of the polymer chain.) Once both probe and target DNA are single-stranded, mixing of the two will allow complementary bases to reassociate. This process is called DNA annealing or reassociation.
|
| A single-stranded probe and target can potentially anneal in a variety of ways:
|
- formation of probe-probe complementary strands (when using a ds probe);
- reannealing of the complementary strands of the target, so-called homoduplexes;
- formation of probe-target heteroduplexes.
|
| page 473 |  | | page 474 |
|
Table 34-1.
Characteristics of some nucleic acid probes used for hybridization studies. |
| Body_ID: None |
| Nucleic acid probes used for hybridization studies |
| Body_ID: T034001.50 |
| Probe type | Origin | Probe characteristics | Labeling method |
| Body_ID: T034001.100 |
| DNA | cell-based DNA: cloning, | double-stranded | random primer |
| Body_ID: T034001.150 |
| | polymerase chain | cell-based: 0.1-100+kb | nick translation |
| Body_ID: T034001.200 |
| | reaction (PCR) | PCR-based: 0.1-10 kb | |
| Body_ID: T034001.250 |
| RNA | RNA transcription from phage vectors | single stranded: 1-2 kb | run-off transcription |
| Body_ID: T034001.300 |
| Oligonucleotide | chemical synthesis | single-stranded: 15-50 nucleotides | end-labeling |
| Body_ID: T034001.350 |
| Formation of probe-target heteroduplexes is the key to the usefulness of molecular hybridization
|
| The conditions under which DNA hybridization occurs and the reliability and specificity, or stringency, of hybridization are affected by several factors:
|
- base composition: GC pairs have three hydrogen bonds compared with the two in an AT pair. Double-stranded DNA with a high GC content is therefore more resistant to denaturation and melts at a higher temperature;
- strand length: the longer a strand of DNA, the greater the number of hydrogen bonds between the two strands. Longer strands require higher temperatures or stronger alkali treatment to denature them; stability varies dramatically with length for very short probes, but above a few hundred base pairs, stability is relatively insensitive to length and is determined primarily by base composition;
- reaction conditions: if the sodium concentration of the reaction mix is high, then the formation of double-stranded DNA is favored, whereas substances that disrupt hydrogen bonds in DNA, (e.g. urea
 or formamide), favor single-stranded DNA. or formamide), favor single-stranded DNA.
|
| Thus if a probe is small, e.g. 50 bp, then when stringency is high, i.e. if the probe and template form in the presence of moderate sodium concentration and at high temperature, a single base pair difference may prevent the formation of the probe - target duplex, whereas a larger probe, e.g. 500 bp, would still form a stable duplex. The converse may also be true, i.e. if there are substantial differences between the probe and target, annealing of the probe and target may occur even under conditions of low stringency (high sodium and low temperature) (Fig. 34.1).
|
| Figure 34.1 Probe-template hybridization. (A) Large probes, e.g. 200 bases or more, can form stable heteroduplexes with the target DNA even if there are a significant number of non-complementary bases in conditions of low stringency. (B) Oligonucleotide probes, in contrast, may discriminate between targets that differ by a single base under stringent conditions. |
| page 474 |  | | page 475 |
| One means of measuring the stability of a nucleic acid duplex is assessing its melting temperature (Tm)
|
The melting temperature (Tm) is the temperature, in vitro, at which 50% of a double-stranded duplex has dissociated into single-strand form. There are several ways of determining Tm but one simple formula for estimating the melting temperature of moderately sized oligonucleotides, is as follows:
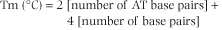
|
| This formula emphasizes the importance of the number of hydrogen bonds present between DNA strands and the energy required to disrupt double-stranded DNA. The Tm for human DNA is approximately 87°C under standard conditions and reflects the average GC content - about 40% - of human DNA.
|
| Probes must have a label to be identified
|
| Implicit in the use of probes to identify pieces of complementary DNA is the notion that if hybridization occurs, the heteroduplex can be specifically detected. The probe is labeled so that the probe-target duplex can be identified. There are many ways in which probes can be labeled, but they fall into two categories, either isotopic, i.e. involving radioactive atoms, or nonisotopic, e.g. end-labeling probes with fluorescent tags or small ligand molecules (Table 34.2).
|
| The most commonly used imaging technique is direct autoradiography, which involves placing the sample in direct contact with the photographic material, usually an X-ray film. The radioactivity of the bound probe produces a dark image on the developed film. 32P is a commonly used label for nucleic acid probes because of its high specific activity. The beta particle emitted is sufficiently energetic that a substantial fraction passes through ordinary film without exposing it. Therefore it is common to use the same type of intensifying screens used clinically to make film more sensitive to X-rays. Energetic beta particles pass through the film but are absorbed by the intensifying screen, which then emits visible light and exposes the film. |
| The majority of techniques involving probe hybridization and labeling still involve the use of radioisotopes such as 32P, 35S or 3H and, as such, require a method for detecting and localizing the radioactivity. The most common method involves the process of autoradiography. Autoradiography allows information from a solid phase, e.g. as a gel or fixed-tissue
samples, to be saved in two-dimensional form as an exposed photographic image.
|
|