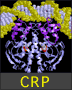
| The sidechains of Tyr13, Phe56, and Asp102 are shown as Spacefill with CPK coloring. The bases of Cyt 10, Ade 11, and Cyt 12 are shown as Spacefill with Shapely coloring. Tyr13 stacks atop Cyt 10 and forms a perpendicular "aromatic-aromatic" interaction with Phe56. Phe56 and Asp102 sandwich the bases of Ade 11 and Cyt 12 in a four-member stack. (cf. Fig. 5e of Oubridge et al.)
Viewing tips: Use the buttons to the right of the image to show only portions of the structure; then use the Chime menu to display or color the molecule. The rotate buttons act on the entire structure. Remove the distance monitors with a second click. |